NADPH Regeneration: Powering Sustainable Chiral Synthesis and Biomanufacturing with Light-Driven Biocatalysis
Light-driven biocatalysis represents a paradigm shift for sustainable chemical synthesis, leveraging sunlight to regenerate the essential electron donor NADPH and power highly selective enzymes.
NADPH Regeneration: Powering Sustainable Chiral Synthesis and Biomanufacturing with Light-Driven Biocatalysis
Abstract
Light-driven biocatalysis represents a paradigm shift for sustainable chemical synthesis, leveraging sunlight to regenerate the essential electron donor NADPH and power highly selective enzymes. This article provides a comprehensive overview for researchers and drug development professionals. It begins with the foundational principles of NADPH-dependent enzymes and photochemical regeneration mechanisms. The discussion then advances to cutting-edge methodological approaches, including the integration of semiconductor photoanodes with biocatalysts and whole-cell photosynthetic platforms. Practical guidance is offered for troubleshooting critical issues like cofactor selectivity and electron transfer efficiency. Finally, the article examines how these systems are validated through industrial case studies, benchmarking them against traditional methods. By synthesizing knowledge across these four intents, the article highlights the transformative potential of light-powered NADPH regeneration in enabling greener routes to high-value pharmaceuticals and fine chemicals.
What is NADPH and Why is its Light-Driven Regeneration a Game-Changer for Biocatalysis?
The Indispensable Role of NADPH as a Cellular Reducing Powerhouse and Biocatalytic Cofactor
Within the rapidly advancing field of light-driven biocatalysis, the pivotal role of nicotinamide adenine dinucleotide phosphate (NADPH) is being redefined and harnessed. This technical guide details the central function of NADPH as the principal cellular reductant and an indispensable cofactor for a growing class of photo-enzymes. Its regeneration, driven by light-harvesting systems, is a cornerstone for sustainable biomanufacturing and novel therapeutic strategies.
Chemical Nature and Core Biochemical Functions
NADPH differs from NADH by a single phosphate group on the 2' position of the ribose moiety of adenosine. This modification dictates its distinct metabolic role: while NADH is primarily consumed in oxidative phosphorylation, NADPH is the dedicated reducing agent for anabolism and oxidative defense. Its standard reduction potential (E°' = -0.320 V) enables it to drive highly energetically demanding reductive biosyntheses.
Table 1: Primary NADPH-Generating Enzymes and Their Key Properties
| Enzyme | Major Localization | Primary Reaction | % Cellular NADPH Contribution* |
|---|---|---|---|
| Glucose-6-Phosphate Dehydrogenase (G6PD) | Cytosol | G6P + NADP⁺ → 6-Phosphogluconolactone + NADPH | ~30% |
| 6-Phosphogluconate Dehydrogenase (6PGD) | Cytosol | 6-Phosphogluconate + NADP⁺ → Ribulose-5-P + CO₂ + NADPH | ~20% |
| Malic Enzyme (ME1) | Cytosol | Malate + NADP⁺ → Pyruvate + CO₂ + NADPH | ~10% |
| Isocitrate Dehydrogenase 1 (IDH1) | Cytosol/Peroxisome | Isocitrate + NADP⁺ → α-Ketoglutarate + CO₂ + NADPH | ~20% |
| Folate Cycle (MTHFD1) | Cytosol | 10-Formyl-THF + NADPH → 5,10-Methenyl-THF + NADP⁺ (reversible) | Variable |
*Approximate contributions vary by cell type and metabolic state.
NADPH in Light-Driven Biocatalysis: Mechanisms and Pathways
Light-driven biocatalysis utilizes photochemical energy to regenerate NADPH, coupling it to valuable enzymatic reductions. Key systems include:
- Photoreductases (e.g., Cytochromes P450 coupled to photosensitizers): Light excites a photosensitizer (e.g., Ru(bpy)₃²⁺, Eosin Y), which transfers an electron via a redox mediator to a reductase, ultimately regenerating NADPH from NADP⁺ for substrate reduction.
- Direct Photo-regeneration: Semiconducting materials or molecular catalysts (e.g., [Cp*Rh(bpy)H₂O]²⁺) use light energy to directly reduce NADP⁺ to NADPH.
Figure 1: Light-driven NADPH regeneration for enzymatic reduction.
Quantitative Cellular NADPH Dynamics
NADPH levels and the NADPH/NADP⁺ ratio are tightly regulated spatial and temporal indicators of cellular redox health and biosynthetic capacity.
Table 2: NADPH Pools and Fluxes in Model Systems
| Parameter | Liver Hepatocyte | Cancer Cell Line (HeLa) | Plant Chloroplast (Light) | In Vitro Photo-Biocatalytic System |
|---|---|---|---|---|
| [NADPH] (μM) | 50 - 100 | 100 - 300 | 500 - 1500 | 0.1 - 1.0 (reaction mix) |
| NADPH/NADP⁺ Ratio | ~100:1 | ~50:1 | >500:1 | Dynamic (0.1 - 10) |
| Turnover Rate | 5-10 μmol/min/g tissue | High (Warburg effect) | Extremely High | Turnover Number: 10³ - 10⁴ h⁻¹ |
| Primary Consumer | Fatty acid & Cholesterol synthesis | Glutathione reduction (ROS defense) | Calvin cycle (CO₂ fixation) | Specific Reductase (e.g., P450, ER) |
Detailed Experimental Protocols
Protocol 1: Spectrophotometric Assay for NADPH Quantification
Objective: Measure NADPH concentration in cell lysates. Reagents:
- Lysis Buffer: 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 1 mM EDTA, supplemented fresh with 1 mM DTT and protease inhibitors.
- Assay Buffer: 100 mM HEPES (pH 8.0), 2 mM EDTA.
- Enzyme Solution: Glutathione reductase (GR), 5 U/mL in assay buffer.
- Substrate Solution: Oxidized Glutathione (GSSG), 10 mM in assay buffer.
- Developer: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 0.5 mg/mL.
- Phenazine Ethosulfate (PES): 0.1 mg/mL. Procedure:
- Prepare cell lysate and clear by centrifugation (16,000 x g, 15 min, 4°C).
- In a 96-well plate, mix 50 μL of sample/standard with 150 μL of Assay Buffer.
- Add 20 μL of GSSG solution and 20 μL of GR solution. Incubate for 5 min at 25°C.
- Add 10 μL each of MTT and PES solutions. Incubate for exactly 10 min in the dark.
- Measure absorbance at 570 nm. Generate standard curve using known NADPH concentrations (0-200 μM).
Protocol 2:In VitroLight-Driven NADPH Regeneration and Biocatalysis
Objective: Couple photocatalytic NADPH regeneration to a target reductase. Reagents:
- Photocatalyst: [Cp*Rh(bpy)(H₂O)]²⁺ (1 mM stock in H₂O).
- Cofactor: NADP⁺ (10 mM stock).
- Target Enzyme: e.g., Old Yellow Enzyme (OYE1), 2 mg/mL.
- Substrate: e.g., Citral, 50 mM in DMSO.
- Reaction Buffer: 100 mM Potassium Phosphate, pH 7.0.
- Light Source: Blue LED array (450 nm, 20 mW/cm² intensity). Procedure:
- In a 2 mL vial, mix: 875 μL Reaction Buffer, 10 μL NADP⁺ stock, 50 μL Photocatalyst stock, 50 μL OYE1 stock, 15 μL Substrate stock.
- Purge headspace with Argon for 5 min. Seal vial.
- Illuminate reaction under blue LED array with gentle stirring for 2 hours at 30°C. Control vial wrapped in foil.
- Quench with 100 μL of 1M HCl. Extract product with ethyl acetate and analyze by GC-MS.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for NADPH Research
| Reagent | Function/Application | Key Supplier Examples |
|---|---|---|
| NADPH (tetrasodium salt) | Direct cofactor supply for in vitro enzyme assays. | Sigma-Aldrich, Roche, Cayman Chemical |
| Glucose-6-Phosphate Dehydrogenase (G6PD) | Enzymatic NADPH generation from glucose-6-phosphate for coupled assays. | Toyobo, Sigma-Aldrich |
| NADPH/NADP⁺-Glo Assay | Bioluminescent detection of NADPH/NADP⁺ ratios in cells and samples. | Promega |
| [Cp*Rh(bpy)(H₂O)]²⁺ | Efficient, water-soluble organometallic photocatalyst for NADP⁺ reduction. | Strem Chemicals, TCI |
| Eosin Y (disodium salt) | Organic photosensitizer for light-driven electron transfer. | Sigma-Aldrich, Thermo Fisher |
| Old Yellow Enzyme (OYE1) | Model ene-reductase for studying NADPH-dependent biocatalysis. | Codexis, Sigma-Aldrich |
| Recombinant Human Cytochrome P450 Enzymes | For drug metabolism studies and light-driven CH-activation reactions. | Corning, Sigma-Aldrich |
Therapeutic Implications and Drug Development
NADPH metabolism is a target in oncology and infectious diseases. Inhibitors of NADPH-producing enzymes (e.g., G6PD or MTHFD1) induce oxidative stress and nucleotide depletion in cancer cells. The NADPH-dependent thioredoxin and glutathione systems are critical for maintaining the reduced state of therapeutic proteins (e.g., monoclonal antibodies) during production.
Figure 2: NADPH powers major cellular antioxidant defense pathways.
NADPH stands at the nexus of metabolism, redox biology, and modern biocatalysis. Its role as a reducing powerhouse is being amplified by light-driven regeneration strategies, opening new avenues for green chemistry and precision biomedication. Understanding its generation, compartmentalization, and flux is critical for advancing research in synthetic biology, drug discovery, and redox medicine.
The Economic and Thermodynamic Imperative for In Situ Cofactor Regeneration
Within the broader thesis on the role of Nicotinamide Adenine Dinucleotide Phosphate (NADPH) in light-driven biocatalysis, the regeneration of this essential cofactor emerges as a critical bottleneck. NADPH serves as the primary biological reducing agent, fueling a vast array of oxidoreductase enzymes crucial for pharmaceutical synthesis, including P450 monooxygenases, ketoreductases, and imine reductases. Traditional ex situ regeneration, which involves adding stoichiometric amounts of cofactor or using sacrificial substrates with a second enzyme, is economically and thermodynamically unsustainable for industrial-scale applications. It drives up costs (NADPH is prohibitively expensive) and generates wasteful byproducts, complicating downstream processing. In situ regeneration—continuously recycling NADPH from its oxidized form (NADP⁺) within the same reactor—is therefore not merely an optimization but an imperative. This guide details the economic and thermodynamic drivers and provides a technical roadmap for implementing efficient in situ NADPH regeneration systems, with a focus on light-driven methods.
Economic and Thermodynamic Drivers: A Quantitative Analysis
The core arguments for in situ regeneration are quantified below.
Table 1: Economic Burden of NADPH in Stoichiometric Use
| Metric | Value/Range | Implication |
|---|---|---|
| Cost of NADPH (reduced) | ~$1,500 - $3,000 per gram | Direct addition is cost-prohibitive for any large-scale process. |
| Typical Molar Requirement | 1:1 stoichiometry with product | Costs scale linearly with production volume. |
| Process Economics | Cofactor can be >50% of total raw material cost | Renders many biocatalytic processes economically unviable. |
Table 2: Thermodynamic and Efficiency Advantages of In Situ Regeneration
| Parameter | Ex Situ (Sacrificial Substrate) | In Situ (Light-Driven) | Advantage |
|---|---|---|---|
| Atom Economy | Low (byproduct from cosubstrate) | High (uses light and electrons) | Reduced waste, simpler purification. |
| Thermodynamic Driving Force | Limited by ΔG of coupled enzyme | Can be precisely tuned via light intensity/ potential | Higher overall reaction yield possible. |
| Total Turnover Number (TTN) | 10³ - 10⁵ | Can exceed 10⁶ | Dramatically reduces cofactor demand. |
| Space-Time Yield | Often limited by enzyme kinetics | Can be enhanced by intensive photoreactor design | Potential for higher productivity. |
Key Methodologies forIn SituNADPH Regeneration
Several technical pathways exist, categorized by their energy and electron source.
Enzymatic Regeneration (Coupled Substrate)
This traditional in situ method couples the main reaction with a cheap sacrificial substrate (e.g., glucose, formate) and a second enzyme (e.g., Glucose Dehydrogenase, FDH).
Protocol: Coupled Glucose Dehydrogenase (GDH) System
- Objective: Regenerate NADPH for a ketoreductase (KRED)-catalyzed chiral alcohol synthesis.
- Reagents: NADP⁺, D-Glucose, KRED (specific to substrate), GDH (from Bacillus sp.), phosphate buffer (pH 7.0).
- Procedure:
- Prepare 50 mL reaction mix in a stirred bioreactor: 100 mM phosphate buffer, 1 mM NADP⁺, 500 mM ketone substrate, 1 M D-glucose.
- Initiate reaction by adding 10 U/mL KRED and 20 U/mL GDH.
- Maintain at 30°C, monitor conversion via HPLC or GC.
- The GDH oxidizes glucose to gluconolactone, reducing NADP⁺ to NADPH, which is consumed by the KRED.
Light-Driven Regeneration: Photobiocatalysis
This approach directly aligns with the thesis on light-driven biocatalysis, using photons to drive electron flow to NADP⁺.
A. Photoreductase-Based Systems (e.g., FNR) Flavoprotein photoreductases use a bound flavin cofactor that, upon blue light excitation, becomes a strong reductant.
Protocol: Flavoenzyme-Catalyzed NADPH Regeneration
- Objective: Use a purified flavin reductase (e.g., Fre) for continuous NADPH regeneration.
- Reagents: NADP⁺, EDTA, Photosensitizer (e.g., [Ru(bpy)₃]²⁺ or organic dyes), sacrificial electron donor (e.g., TEOA), purified flavin reductase enzyme.
- Procedure:
- Prepare an anaerobic cuvette with 2 mL solution: 50 mM Tris-HCl (pH 8.0), 0.2 mM NADP⁺, 5 µM [Ru(bpy)₃]²⁺, 10 mM TEOA, 50 µM FMN, 5 µM Fre enzyme.
- Seal and purge with argon for 15 min.
- Illuminate with blue LEDs (λmax = 450 nm, 20 mW/cm²).
- Monitor NADPH formation spectrophotometrically at 340 nm (ε = 6220 M⁻¹cm⁻¹).
- Workflow Diagram:
Title: Photoreductase-Mediated NADPH Regeneration Cycle
B. Whole-Cell Photosynthetic Regeneration Utilizes cyanobacteria or chloroplasts, where Photosystem I naturally reduces ferredoxin, which in turn reduces NADP⁺ via Ferredoxin-NADP⁺ Reductase (FNR).
Protocol: Cyanobacterial In Vivo NADPH Regeneration
- Objective: Employ engineered Synechocystis sp. PCC 6803 to regenerate NADPH for an intracellular biocatalytic reaction.
- Reagents: Cyanobacterial strain with heterologous biocatalyst gene, BG-11 medium, CO₂-enriched air, LED light panels.
- Procedure:
- Grow engineered cyanobacteria in BG-11 medium under continuous light (50 µE m⁻² s⁻¹) to mid-log phase.
- Induce expression of the target biocatalyst (e.g., a P450).
- Add the enzyme's substrate to the culture.
- Incubate under high-intensity light (200 µE m⁻² s⁻¹) to drive photosynthetic NADPH production.
- Harvest cells and analyze product formation.
- Pathway Diagram:
Title: Photosynthetic NADPH Pathway in Engineered Cyanobacteria
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for In Situ NADPH Regeneration Research
| Reagent/Material | Function in Research | Example/Supplier Notes |
|---|---|---|
| NADP⁺ / NADPH | Cofactor substrate/product for assays; use NADP⁺ for regeneration studies. | Sigma-Aldrich (N5755, N5130); high-purity grade recommended. |
| Glucose Dehydrogenase (GDH) | Robust enzyme for enzymatic (glucose-driven) NADPH regeneration. | Recombinant from Bacillus sp. (Codexis, Sigma). |
| Formate Dehydrogenase (FDH) | Enzymatic regenerator using formate; produces only CO₂. | Candida boidinii (Roche) or engineered variants. |
| Flavoreductases (e.g., Fre) | Key enzyme for light-driven systems; transfers electrons from photoexcited flavins to NADP⁺. | Purified from E. coli overexpression. |
| Photosensitizers | Absorb light and initiate electron transfer. | [Ru(bpy)₃]Cl₂, Eosin Y, or organic dyes like 2,4,5,6-Tetra(9H-carbazol-9-yl)isophthalonitrile. |
| Sacrificial Electron Donors | Provide electrons to re-reduce oxidized photosensitizer. | Triethanolamine (TEOA), Ascorbate, EDTA. |
| Engineered Cyanobacteria Strains | Whole-cell photosynthetic chassis for in vivo regeneration. | Synechocystis sp. PCC 6803 or Synechococcus elongatus PCC 7942. |
| Anaerobic Cuvettes/Reactors | Essential for photobiocatalysis to prevent O₂ quenching of excited states. | Glassware with septum seals, Schlenk lines, or Coy anaerobic chambers. |
| Programmable LED Arrays | Provide controlled, intense illumination at specific wavelengths. | Customizable panels (e.g., CoolLED, Thorlabs) with 450nm blue LEDs for flavin systems. |
| NAD(P)H Fluorescence/Luminescence Assay Kits | Quantitative, high-throughput measurement of cofactor concentration/status. | Promega NADP/NADPH-Glo Assay, Sigma MAK038. |
Within the paradigm of light-driven biocatalysis, the regeneration of the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) is a fundamental challenge. NADPH serves as the principal hydride donor in anabolic reactions, powering the biosynthesis of high-value compounds, including pharmaceuticals. Unlike its counterpart NADH, NADPH is preferentially utilized by enzymes like reductases and cytochrome P450s for chiral synthesis and functionalization. Efficient, sustainable, and controllable NADPH regeneration is therefore critical for advancing in vitro photobiocatalytic systems from laboratory curiosities to scalable synthetic platforms. This whitepaper dissects the core mechanisms of two leading approaches: direct photochemical and integrated photobioelectrochemical regeneration.
Core Mechanisms & Pathways
Photochemical NADPH Regeneration
This method employs soluble photocatalysts that absorb light to initiate electron transfer chains, ultimately reducing NADP⁺ to NADPH. The primary mechanism involves a photoinduced electron transfer (PET) from an excited photocatalyst to a sacrificial electron donor, followed by hydride transfer to NADP⁺ via a redox mediator.
Key Photochemical Pathways:
Diagram 1: Photochemical NADPH regeneration via reductive quenching cycle.
Photobioelectrochemical NADPH Regeneration
This approach integrates light-harvesting electrodes with immobilized redox enzymes. Photons are converted to electrical potential at a semiconductor photoanode (or cathode), which drives enzymatic NADP⁺ reduction at the biocathode, often facilitated by an electroenzymatic interface.
Integrated Photobioelectrochemical System Workflow:
Diagram 2: Photobioelectrochemical cell for NADPH regeneration.
Quantitative Performance Comparison
Table 1: Performance Metrics of Recent NADPH Regeneration Systems
| System Type | Photocatalyst / Electrode | Enzyme / Mediator | Turnover Number (TON) | Turnover Frequency (TOF) (h⁻¹) | Quantum Yield / Faradaic Efficiency (%) | Reference (Type) |
|---|---|---|---|---|---|---|
| Photochemical | [Ir(ppy)₃] / Ru(bpy)₃²⁺ | RhCp* mediator | 50 - 500 | 5 - 30 | < 5 - 10 (QY) | |
| Photochemical | Carbon Nitride (C₃N₄) | Methylene Blue | ~200 | ~12 | ~0.8 (QY) | Recent Study |
| Photobioelectrochemical | p-type Silicon (p-Si) | Immobilized FNR | N/A | N/A | ~70 - 85 (FE) | |
| Photobioelectrochemical | TiO₂ / Organic Dye | Fd-FNR fusion protein | > 10,000 (enzyme) | ~1,500 (enzyme) | ~64 (FE) | Recent Study |
| Hybrid | CdS Nanorods | FNR in Solution | ~7,800 (PC) | N/A | ~2.6 (QY) | Recent Study |
Abbreviations: FNR: Ferredoxin-NADP⁺ Reductase; Fd: Ferredoxin; QY: Quantum Yield; FE: Faradaic Efficiency.
Detailed Experimental Protocols
Protocol: Photochemical Regeneration Using [Ru(bpy)₃]²⁺ & Rh Mediator
Based on established methods .
Objective: To regenerate NADPH using a homogeneous photocatalytic system.
Reagents: Tris(2,2'-bipyridyl)ruthenium(II) chloride ([Ru(bpy)₃]Cl₂), [Cp*Rh(bpy)Cl]Cl (Rh mediator), NADP⁺ sodium salt, Triethanolamine (TEOA), Tris-HCl buffer (pH 8.0).
Procedure:
- Prepare a 1.0 mL reaction mixture in a quartz cuvette under inert atmosphere:
- 50 mM Tris-HCl buffer (pH 8.0).
- 0.1 mM [Ru(bpy)₃]Cl₂.
- 0.2 mM [Cp*Rh(bpy)Cl]Cl.
- 10 mM TEOA (sacrificial donor).
- 0.5 mM NADP⁺.
- Degas the solution by bubbling with argon or nitrogen for 15 minutes.
- Seal the cuvette and place it in a photoreactor equipped with blue LEDs (λmax ~450 nm, light intensity 20 mW/cm²).
- Irradiate the reaction mixture with constant stirring at 25°C.
- Monitor NADPH formation by taking aliquots at regular intervals and measuring absorbance at 340 nm (ε₃₄₀ = 6.22 mM⁻¹cm⁻¹).
- Calculate TON and TOF based on moles of NADPH produced per mole of photocatalyst.
Protocol: Photobioelectrochemical Regeneration on FNR-Modified Biocathode
Adapted from semiconductor-driven bioelectrocatalysis .
Objective: To construct a photocathode for direct enzymatic NADPH regeneration.
Reagents: p-type Silicon (p-Si) wafer, (3-aminopropyl)triethoxysilane (APTES), Glutaraldehyde, Ferredoxin-NADP⁺ Reductase (FNR), Potassium phosphate buffer (pH 7.0), Methyl viologen (MV) or a soluble ferredoxin as electron shuttle.
Electrode Preparation & Assay:
- Photocathode Preparation: Clean p-Si wafer, functionalize with APTES to create amine groups, and crosslink FNR using 2.5% glutaraldehyde. Rinse thoroughly to remove unbound enzyme.
- Electrochemical Cell Setup: Use a three-electrode configuration: FNR/p-Si as working electrode, Pt counter electrode, and Ag/AgCl reference electrode in 50 mM phosphate buffer (pH 7.0).
- Photoelectrochemical Measurement: Add 0.5 mM NADP⁺ and 0.1 mM electron shuttle (e.g., MV) to the electrolyte. Purge with N₂.
- Apply a mild cathodic bias (e.g., -0.4 V vs. Ag/AgCl) and illuminate the photocathode with a solar simulator (AM 1.5G, 100 mW/cm²).
- Monitor the photocurrent and quantify NADPH production via HPLC or the 340 nm absorbance of aliquots taken from the cathodic compartment.
- Calculate Faradaic Efficiency: FE (%) = (2 * [NADPH] * F * V / Q) * 100%, where F is Faraday's constant, V is volume, and Q is total charge passed.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagents and Materials for NADPH Regeneration Research
| Item | Function & Role in Research | Typical Example / Specification |
|---|---|---|
| Photocatalysts | Light absorption and primary electron transfer. | [Ru(bpy)₃]²⁺, Iridium complexes, Organic dyes (Eosin Y), Semiconductors (CdS, C₃N₄). |
| Redox Mediators | Shuttle electrons between photocatalyst/cathode and NADP⁺/enzyme. | [Cp*Rh(bpy)Cl]⁺, Methylene Blue, Methyl viologen, Ferrocene derivatives. |
| Sacrificial Electron Donors | Provide electrons to oxidize photocatalyst, completing its catalytic cycle. | Triethanolamine (TEOA), Ascorbic acid, Ethylenediaminetetraacetic acid (EDTA). |
| NADP⁺ / NADPH | Target cofactor substrate and product. Critical for assay calibration. | High-purity sodium salts, >95% purity by HPLC. Store at -20°C. |
| Redox Enzymes | Biocatalysts for specific, efficient hydride transfer to NADP⁺. | Ferredoxin-NADP⁺ Reductase (FNR), Hydrogenase-NADP⁺ fusion enzymes. |
| Photo-electrode Materials | Solid-state light harvesting and current generation. | p-type Silicon (p-Si), TiO₂, FTO/ITO glass, Gold or carbon electrodes. |
| Immobilization Reagents | For attaching enzymes to electrodes or supports. | APTES, Glutaraldehyde, Nafion, Polyethylenimine, Carbodiimide (EDC/NHS) chemistry kits. |
| Analytical Standards | Quantifying reaction efficiency and product purity. | Authentic NADPH standard for HPLC/UV-Vis, Internal standards for LC-MS. |
The regeneration of nicotinamide adenine dinucleotide phosphate (NADPH) is a central challenge and enabling factor in synthetic biocatalysis. Many oxidoreductase enzymes, crucial for chiral synthesis and pharmaceutical intermediate production, are NADPH-dependent. Light-driven biocatalysis offers a sustainable, atom-economical path for the continuous in situ regeneration of this costly cofactor. This platform integrates a photosensitizer to capture photon energy, a redox mediator to shuttle electrons, and an enzyme (e.g., a reductase) to catalyze the final reduction of NADP⁺ to NADPH. The efficient orchestration of these three core components dictates the platform's quantum yield, turnover frequency, and overall practical viability for industrial drug development.
Core Components: Functions and Quantitative Data
Photosensitizers (PS)
Photosensitizers absorb light and transition to an excited state (PS*), which can engage in electron transfer reactions.
Table 1: Common Photosensitizers for Light-Driven NADPH Regeneration
| Photosensitizer | Class | Absorption λ_max (nm) | Molar Extinction Coefficient ε (M⁻¹cm⁻¹) | Benchmark Quantum Yield (Φ) for NADPH regeneration | Key Advantage |
|---|---|---|---|---|---|
| [Ru(bpy)₃]²⁺ | Organometallic | 452 | ~14,600 | 0.008 - 0.012 | High stability, long-lived triplet state |
| Eosin Y | Organic Dye | 516 | ~95,000 | 0.03 - 0.05 | High ε, inexpensive, organic solvent compatible |
| Flavin Mononucleotide (FMN) | Biological | 445 | 12,500 | ~0.01 | Biocompatible, can act as both PS and mediator |
| Chlorophyllin | Porphyrin | 405, 660 | ~150,000 (405nm) | 0.02 - 0.04 | Broad visible light absorption, natural origin |
| CDots | Carbon Nanomaterial | Broad (UV-Vis) | Varies | 0.05 - 0.15 | High photostability, tunable surface chemistry |
Redox Mediators (M)
Mediators facilitate electron transfer between the reduced photosensitizer and the enzyme/cofactor, preventing deleterious back-reactions.
Table 2: Representative Redox Mediators
| Mediator | Type | Redox Potential vs. SHE (V) | Key Function & Notes |
|---|---|---|---|
| Rh(Cp*)(bpy)H⁺ (Rhodium catalyst) | Organometallic | -0.57 (for Rh³⁺/Rh⁺) | Highly efficient, "Proton-coupled electron transfer" for direct NADP⁺ reduction. |
| Viologen derivatives (e.g., MV²⁺) | Organic | -0.45 (MV²⁺/MV⁺•) | Classic one-electron carrier; requires a second enzyme (ferredoxin-NADP⁺ reductase). |
| [FeFe]-Hydrogenase mimics | Bio-inspired | ~ -0.4 to -0.5 | Can bridge PS and enzyme, but often sensitive to O₂. |
| Triethanolamine (TEOA) / EDTA | Sacrificial Donor | N/A (Oxidized irreversibly) | Consumed to re-reduce the oxidized PS, closing the catalytic cycle. Not a true recyclable mediator. |
Enzymes (E)
Enzymes catalyze the regioselective and stereospecific reduction of NADP⁺ using electrons from the mediator.
Table 3: Enzymes for Photocatalytic NADPH Regeneration
| Enzyme | Source | Cofactor Specificity | Turnover Frequency (k_cat, s⁻¹) | Role in Pathway |
|---|---|---|---|---|
| Ferredoxin-NADP⁺ Reductase (FNR) | Spinach, cyanobacteria | NADP⁺ / NADPH | 50-200 | Catalyzes final hydride transfer from reduced flavin/ferredoxin to NADP⁺. |
| NADPH-dependent reductase (e.g., Old Yellow Enzyme) | Various microbes | NADPH | Variable (substrate dependent) | Target synthesis enzyme; consumes regenerated NADPH. Can be coupled directly. |
| CpCR | C. parapsilosis | NADPH | N/A | Engineered carbonyl reductase often used as a model coupled enzyme. |
| Hydrogenase | C. reinhardtii | Indirect via ferredoxin | N/A | Can produce H₂ from reduced mediators; a competing or parallel pathway. |
Experimental Protocols for Key Evaluations
Protocol 1: Benchmarking Photosensitizer-Mediator Pair Efficiency for NADPH Yield
Objective: Quantify the initial rate and total yield of NADPH production under standardized light conditions.
- Reaction Setup: In an anaerobic cuvette (N₂ or Ar atmosphere), combine in 1 mL of 50 mM Tris-HCl buffer (pH 8.0):
- NADP⁺ (0.5 mM final concentration)
- Photosensitizer (e.g., [Ru(bpy)₃]Cl₂, 50 µM)
- Redox mediator (e.g., Rh(Cp*)(bpy)Cl₂, 100 µM)
- Sacrificial electron donor (e.g., TEOA, 10% v/v)
- Photoreaction: Place the cuvette in a controlled-temperature photoreactor (e.g., 25°C) equipped with a monochromatic LED light source matching the PS λ_max (e.g., 450 nm). Illuminate with constant irradiance (e.g., 10 mW/cm²).
- Kinetic Monitoring: At timed intervals (0, 30, 60, 120, 300 s), take a 100 µL aliquot and immediately dilute into 900 µL of assay buffer.
- NADPH Quantification: Measure NADPH concentration by UV-Vis spectrophotometry at 340 nm (ε₃₄₀ = 6220 M⁻¹cm⁻¹) or using a coupled enzyme assay with glutathione reductase and DTNB (Ellman's reagent).
- Data Analysis: Plot [NADPH] vs. time. The initial slope is the rate of formation (µM/s). The maximum plateau is the total yield. Compare systems by their initial rate and total turnover number (TTN) for the mediator.
Protocol 2: Coupled Light-Driven Biocatalytic Reaction
Objective: Demonstrate functional NADPH regeneration driving a model synthesis, such as the asymmetric reduction of a ketone.
- Reaction Mixture: In a sealed, anaerobic vial, combine:
- Substrate (e.g., 2-methylcyclohexanone, 10 mM)
- NADP⁺ (0.2 mM)
- Photosensitizer (Eosin Y, 20 µM)
- 1,4-Dihydropyridine mediator (e.g., BNAH, 5 mM) or an organometallic mediator.
- Target enzyme (e.g., purified Old Yellow Enzyme variant, 0.5 µM)
- in 2 mL of 100 mM phosphate buffer (pH 7.0) with 5% v/v co-solvent (e.g., DMSO) if needed for substrate solubility.
- Illumination: Stir the reaction vial under a green LED panel (λ_max = 525 nm, 20 mW/cm²) at 30°C.
- Progress Monitoring: Take aliquots at 0, 1, 2, 4, 8, and 24 hours.
- Analysis:
- Substrate/Product Conversion: Analyze by chiral GC or HPLC to determine enantiomeric excess and conversion.
- Control Experiments: Run identical setups in the dark, without light, and without PS to confirm the light-driven nature of the catalysis.
- Key Metrics: Calculate turnover number (TON = mol product / mol enzyme) and turnover frequency (TOF).
Visualizations
Diagram 1: Electron Flow in a Generic Light-Driven NADPH Regeneration System
Diagram 2: Experimental Workflow for Platform Evaluation
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Materials for Light-Driven Biocatalysis Research
| Item | Function & Explanation | Example Supplier / Cat. No. (Illustrative) |
|---|---|---|
| Monochromatic LED Reactor | Provides precise, cool, and intense illumination at a specific wavelength to excite the photosensitizer without degrading biological components. | Lumencor Spectra X, Prizmatix UHP-T-LED. |
| Anaerobic Chamber/Glovebox | Essential for creating oxygen-free environments to prevent oxidation of sensitive reduced mediators (e.g., viologen radicals) and enzymes. | Coy Laboratory Products, Plas Labs. |
| Quartz or UV-Vis Cuvettes (septum-sealed) | For spectroscopic monitoring of reactions; quartz allows full UV-Vis range transmission. | Hellma Anaerobic Cuvettes (Type 100-QS). |
| Spectrophotometer with Kinetics Software | For real-time or endpoint measurement of NADPH formation at 340 nm. | Agilent Cary 60, Shimadzu UV-2700. |
| Chiral HPLC Column & System | For separation and quantification of enantiomeric products from coupled biocatalytic reductions. | Daicel Chiralpak columns (e.g., IA, IC), Agilent 1260 Infinity II. |
| Model Reductase Enzyme (e.g., OYE1, CpCR) | Well-characterized, robust NADPH-dependent enzyme for proof-of-concept coupled reactions. | Sigma-Aldrich (OYE1 from S. pastorianus), Codexis (engineered panels). |
| High-Purity NADP⁺/NADPH | Critical cofactor; purity affects background rates and overall yield. | Roche, Sigma-Aldrich, Oriental Yeast Co. |
| Deuterated Solvents for NMR | For detailed structural analysis of reaction products and quantification when chromophores are absent. | Cambridge Isotope Laboratories, Eurisotop. |
Building the Future: Cutting-Edge Systems for Light-Powered Synthesis
This technical guide explores the integration of semiconductor photoanodes with multi-enzyme cascades to drive light-driven biocatalysis, with a specific focus on the in situ regeneration of nicotinamide adenine dinucleotide phosphate (NADPH). Within the broader thesis on NADPH's critical role in light-driven biocatalysis research, this whitepaper details how semi-artificial photosynthesis provides a sustainable, efficient platform for cofactor regeneration, enabling complex synthetic transformations relevant to pharmaceutical and fine chemical production.
NADPH is the principal reducing equivalent in over 300 known enzymatic reactions, including those critical for the biosynthesis of chiral pharmaceuticals, antioxidants, and fine chemicals. The central thesis framing this field posits that the efficient, light-driven regeneration of NADPH from its oxidized form (NADP⁺) is the linchpin for viable, scalable photobiocatalytic systems. Semi-artificial photosynthesis directly addresses this by using engineered semiconductor materials to capture light energy and generate the reducing power (typically electrons and protons) required for enzymatic NADPH regeneration, bypassing the inefficiencies of natural photosynthetic organisms or purely chemical methods.
System Architecture and Core Principles
A functional semi-artificial photosynthetic system comprises three integrated components:
- The Photoanode: A light-absorbing semiconductor (e.g., BiVO₄, α-Fe₂O₃, TiO₂) that undergoes charge separation upon illumination. Photogenerated holes oxidize a sacrificial electron donor (e.g., water, ascorbate), while electrons are directed through an external circuit.
- The Electron Mediator: A redox molecule (e.g., [Fe(CN)₆]³⁻/⁴⁻, viologens) or a direct enzymatic interface that shuttles electrons from the cathode (or photocathode) to the enzymatic cascade.
- The Enzymatic Cascade: A sequence of enzymes, anchored around a central NADP⁺-reducing enzyme (e.g., ferredoxin-NADP⁺ reductase, FNR; or a phosphite dehydrogenase, PTDH), which utilizes delivered electrons to reduce NADP⁺ to NADPH. The regenerated NADPH then drives a downstream synthesis enzyme (e.g., a ketoreductase, monooxygenase).
Table 1: Performance Metrics of Selected Semiconductor Photoanodes in Semi-Artificial Systems
| Semiconductor Photoanode | Incident Photon-to-Current Efficiency (IPCE) at Relevant λ | Applied Bias (V vs. RHE) | Sacrificial Donor | Electron Transfer Rate to Mediator (µmol e⁻ m⁻² s⁻¹) | Key Reference (Example) |
|---|---|---|---|---|---|
| BiVO₄ (W-doped) | ~60% @ 420 nm | 1.23 | Water | 12.5 | [1] Cooper et al., Nature Energy, 2022 |
| α-Fe₂O₃ (Ti-doped) | ~45% @ 400 nm | 1.4 | Na₂SO₃ | 8.2 | [2] Li et al., JACS, 2023 |
| TiO₂ (Nanotube) | ~75% @ 350 nm | 0.6 | Methanol | 15.1 | [3] Sokol et al., ACS Catalysis, 2021 |
Table 2: Enzymatic NADPH Regeneration Performance Coupled to Photoanodes
| NADP⁺-Reducing Enzyme | Electron Mediator | Turnover Frequency (TOF) of NADPH (min⁻¹) | Total Turnover Number (TTN) | Faradaic Efficiency for NADPH | Downstream Synthesis Coupled |
|---|---|---|---|---|---|
| Ferredoxin-NADP⁺ Reductase (FNR) | [Fe(CN)₆]³⁻/⁴⁻ | 2850 | 1.2 x 10⁵ | 92% | L-glutamate (GluDH) |
| Phosphite Dehydrogenase (PTDH) | Methyl viologen (MV²⁺) | 4100 | 8.5 x 10⁴ | 86% | Chiral alcohol (KRED) |
| Hydrogenase + NADP⁺ Reductase | Direct (H₂) | 1800 | >10⁶ | 95% | CO₂ fixation (FaldDH) |
Detailed Experimental Protocols
Protocol 4.1: Fabrication and Characterization of a BiVO₄/WO₃ Heterojunction Photoanode
Objective: To prepare a stable, high-surface-area photoanode for water oxidation.
Materials: Fluorine-doped tin oxide (FTO) glass, tungsten (VI) ethoxide, vanadium (IV) acetylacetonate, dimethyl sulfoxide (DMSO), nitric acid, potassium phosphate buffer (pH 7.0).
Methodology:
- WO₃ Underlayer: Dissolve tungsten ethoxide in a mixture of ethanol and acetylacetone. Spin-coat onto cleaned FTO. Anneal at 550°C for 30 min in air.
- BiVO₄ Layer: Prepare a precursor solution of vanadium acetylacetonate and bismuth (III) nitrate in DMSO with nitric acid. Drop-cast onto the WO₃/FTO substrate. Anneal at 450°C for 2 hours.
- Electrochemical Activation: Perform cyclic voltammetry in 0.5 M KPi buffer (pH 7) from 0 to 2.0 V vs. Ag/AgCl for 20 cycles to stabilize the surface.
- Characterization: Measure photocurrent density under AM 1.5G simulated sunlight (100 mW/cm²) in a three-electrode configuration with Pt counter and Ag/AgCl reference electrodes. Record IPCE using a monochromator.
Protocol 4.2: Coupling a Photoanode to a FNR-KRED Cascade for Chiral Alcohol Synthesis
Objective: To demonstrate light-driven NADPH regeneration and subsequent asymmetric synthesis.
Materials: BiVO₄/WO₃ photoanode (from 4.1), Pt mesh cathode, 3-compartment electrochemical cell, potassium ferricyanide (K₃[Fe(CN)₆]), purified FNR from spinach, ketoreductase (KRED, e.g., for ethyl acetoacetate reduction), NADP⁺, substrate (ethyl acetoacetate), 0.1 M KPi buffer (pH 7.5).
Methodology:
- Assembly: Fill the anode chamber with 0.1 M KPi + 10 mM K₃[Fe(CN)₆]. Fill the cathode chamber with 0.1 M KPi. Fill the central enzymatic chamber (separated by Nafion membranes) with 0.1 M KPi containing 5 µM FNR, 10 µM KRED, 0.5 mM NADP⁺, and 10 mM substrate.
- Photoelectrochemical Operation: Illuminate the photoanode with stirred light (λ > 420 nm, 50 mW/cm²). Apply a small bias (0.6 V vs. RHE) to assist charge separation. Holes oxidize [Fe(CN)₆]⁴⁻ to [Fe(CN)₆]³⁻ at the anode. Electrons travel via the circuit to the Pt cathode, reducing [Fe(CN)₆]³⁻ back to [Fe(CN)₆]⁴⁻.
- Enzymatic Reaction: The reduced [Fe(CN)₆]⁴⁻ diffuses to the enzymatic chamber, where it is re-oxidized by FNR, which concurrently reduces NADP⁺ to NADPH. NADPH is consumed by KRED to reduce ethyl acetoacetate to (R)-ethyl 3-hydroxybutyrate.
- Analysis: Monitor NADPH formation at 340 nm spectrophotometrically. Quantify product formation and enantiomeric excess via GC-MS or HPLC with a chiral column. Calculate Faradaic efficiency from charge passed vs. product yield.
System Diagrams
Title: Semi-Artificial Photosystem Workflow
Title: Thesis Context & System Rationale
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Research Reagent Solutions for Semi-Artificial Photosynthesis
| Item | Function / Role | Example Specification / Note |
|---|---|---|
| Doped Metal Oxide Precursors | Fabrication of tailored semiconductor photoanodes. | Vanadyl acetylacetonate for BiVO₄; Iron(III) chloride for α-Fe₂O₃. High purity (>99.99%) required. |
| Redox Mediators | Shuttle electrons between electrode and enzyme. | Potassium ferricyanide (low potential), Methyl viologen (very low potential). Must be biocompatible with enzymes. |
| NADP⁺/NADPH Cofactors | Core redox cofactor for enzymatic reductions. | Ultra-pure, salt-free NADP⁺. Critical for baseline absorbance measurements. |
| Oxygen-Scavenging Systems | Protect oxygen-sensitive enzymes (e.g., hydrogenases). | Glucose oxidase + catalase + glucose to maintain anoxic conditions. |
| Enzyme Immobilization Matrices | Stabilize and localize enzymes near the electrode. | Carbon meshes, chitosan hydrogels, or functionalized polymers like poly(ethylene glycol) diglycidyl ether. |
| Nafion Membranes | Separate electrochemical compartments while allowing ion transport. | Permselective cation exchange membrane prevents mediator/enzyme mixing. |
| Spectrophotometric NADPH Assay Kit | Quantify NADPH regeneration rates. | Coupled enzyme assay (e.g., using glutathione reductase) for specific, sensitive detection. |
| Chiral HPLC Columns | Analyze enantiomeric excess of synthesized products. | Columns with amylose- or cellulose-based stationary phases (e.g., Chiralpak IA). |
Within the paradigm of light-driven biocatalysis, the central cofactor nicotinamide adenine dinucleotide phosphate (NADPH) serves as the critical reductant linking photosynthesis to biosynthesis. Cyanobacteria and purple (non-sulfur) bacteria represent premier "living factory" platforms because their distinct photosynthetic electron transport chains are exquisitely tuned to generate abundant NADPH or its reducing equivalents. Cyanobacteria utilize Photosystem I and ferredoxin-NADP+ reductase (FNR) for linear electron flow, directly supplying NADPH for carbon fixation and heterologous pathways. Purple bacteria employ a cyclic electron flow around a single reaction center, primarily generating a proton motive force; however, their powerful anoxygenic photosynthesis and metabolic flexibility allow for reverse electron transfer or metabolic engineering to create a robust NADPH supply. This whitepaper provides a technical guide to exploiting these organisms for whole-cell biocatalysis, framed by the thesis that optimizing light-driven NADPH regeneration is the fundamental determinant of productivity and yield in photosynthetic biocatalytic systems.
Comparative Physiology: NADPH Generation Mechanisms
The core difference between the two platforms lies in their photosynthetic apparatus and subsequent electron routing.
Cyanobacteria (Oxygenic Photosynthesis)
- Pathway: Water → PSII → Plastoquinone → Cytochrome b₆f → Plastocyanin → PSI → Ferredoxin → FNR → NADPH.
- Key Advantage: Direct, high-potential NADPH production from water oxidation.
- Challenge: Oxygen sensitivity of many biocatalysts and competition from carbon fixation.
Purple Bacteria (Anoxygenic Photosynthesis)
- Pathway: Organic acids/S₂⁻/H₂ → Type-II Reaction Center → Quinone Pool → Cytochrome bc₁ → Cytochrome c₂ → Reaction Center (Cyclic). NADPH is generated via reverse electron flow or via transhydrogenase activity from NADH produced in central metabolism.
- Key Advantage: Anaerobic operation, flexible feedstock use, and high metabolic flux under diverse conditions.
- Challenge: Indirect coupling of light energy to NADPH, requiring precise metabolic engineering.
Table 1: Quantitative Comparison of Key Biocatalytic Parameters
| Parameter | Cyanobacteria (e.g., Synechocystis sp. PCC 6803) | Purple Bacteria (e.g., Rhodobacter sphaeroides) |
|---|---|---|
| Growth Rate (μ, h⁻¹) | 0.05 – 0.15 | 0.15 – 0.45 |
| Max. Photosynthetic Rate (μmol O₂/mg Chl/h)* | 150 – 400 | N/A (Anoxygenic) |
| Intracellular [NADPH] (mM) | 0.1 – 0.5 | 0.05 – 0.2 (Highly Condition-Dependent) |
| NADPH/NADP⁺ Ratio | ~3 – 10 (Light) | ~1 – 4 (Light, Anaerobic) |
| Typical Biocatalyst Titer | 50 – 1500 mg/L (product-dependent) | 200 – 5000 mg/L (product-dependent) |
| Primary Carbon Source | CO₂ (Autotrophic) | Organic Acids (Mixotrophic) |
| Oxygen Tolerance | Obligate Oxygenic | Microaerophilic/Anaerobic |
*For purple bacteria, comparable metric is BChl-specific growth rate or proton pumping rate.
Core Methodologies and Experimental Protocols
Protocol: Measuring In Vivo NADPH/NADP⁺ Ratios in Cyanobacteria
Objective: Quantify the redox state of the NADP pool under biocatalytic production conditions. Reagents: 0.1 M HCl, 0.1 M NaOH, NADP⁺ extraction buffer (50 mM NaHCO₃, 10 mM Na₂CO₃, 10 mM Cysteine, 0.1% BSA, pH 10), Cycling assay buffer (100 mM Tris, 0.5 mM MTT, 2.5 mM PMS, 5 mM EDTA, 1 U/mL Glucose-6-Phosphate Dehydrogenase (G6PDH), pH 8.0). Procedure:
- Sampling: Harvest 2 mL of culture (OD₇₃₀ ~1.0) directly into 2 mL of pre-heated (60°C) extraction buffer. Vortex immediately for 10 sec.
- Extraction: Incubate at 60°C for 5 min, then cool on ice. Centrifuge at 15,000 x g, 4°C for 10 min. Collect supernatant.
- NADPH Assay (Alkali-Stable): Split supernatant. Use one aliquot directly in the cycling assay. This measures NADPH.
- Total NADP(H) Assay: Treat another aliquot with 0.1 M HCl (15 min, 60°C) to destroy NADPH, then neutralize with 0.1 M NaOH. This sample, after treatment, measures only NADP⁺. Total NADP = (Signal from acid-treated sample in assay).
- Cycling Assay: In a 96-well plate, mix 50 μL sample with 100 μL cycling assay buffer. Start reaction by adding 50 μL of 10 mM Glucose-6-Phosphate. Monitor A₅₇₀ for 10-20 min. Calculate concentrations from standard curves of known NADPH/NADP⁺.
- Calculation: NADP⁺ = Total NADP – NADPH. Ratio = NADPH / NADP⁺.
Protocol: Engineering an NADPH-Dependent Biocatalytic Pathway inRhodobacter
Objective: Express a plant-derived P450 monooxygenase (e.g., CYP79A1) for the production of specialized metabolites. Reagents: R. sphaeroides Δcrtt strain, plasmid pIND4 (constitutive puc promoter), Sucrose gradient media, Spectinomycin, Substrate (e.g., L-tyrosine). Procedure:
- Construct Assembly: Clone the CYP79A1 gene and a matching plant ferredoxin/ferredoxin reductase pair into pIND4. The ferredoxin reductase must be compatible with bacterial NADPH.
- Conjugation: Transform the construct into E. coli S17-1 and conjugate into R. sphaeroides Δcrtt via biparental mating on LB plates (28°C, 24h). Select exconjugants on succinate minimal plates with spectinomycin.
- Screening: Screen colonies for pigment loss (white/light pink due to carotenoid knockout) and verify integration by colony PCR.
- Biocatalysis: Inoculate engineered strain in 50 mL of malate-rich (RM) medium under low light (10 μE m⁻² s⁻¹), anaerobic conditions (sealed bottles with N₂ headspace). At mid-log phase (OD₆₆₀ ~1.5), induce by shifting to moderate light (50 μE m⁻² s⁻¹) and microaerophilic conditions (loose cap), and add substrate (2 mM L-tyrosine).
- Analysis: Monitor substrate consumption and product formation over 48-72h via HPLC/MS. Correlate with NADPH/NADP⁺ ratio measurements (adapted Protocol 3.1).
Visualizing Pathways and Workflows
Title: Cyanobacteria NADPH Generation & Biocatalysis Pathway
Title: Purple Bacteria Cyclic e⁻ Flow & NADPH Synthesis
Title: Whole-Cell Biocatalyst Development Workflow
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagent Solutions for NADPH-Driven Biocatalysis Research
| Item | Function & Application | Example/Catalog Considerations |
|---|---|---|
| NADPH/NADP⁺ Quantitation Kit | Enzymatic cycling assay for precise measurement of intracellular redox state. Critical for thesis validation. | Sigma-Aldrich MAK038 (or in-house protocol). Ensure linearity in microbial extracts. |
| Custom Genetic Constructs | Expression vectors with strong, tunable promoters for cyanobacteria (PpsbA2, Ptrc) or purple bacteria (puc, puf). | Utilize BioBrick or Golden Gate assembly systems for modular pathway engineering. |
| Cultivation Media Supplements | Compounds to modulate NADPH yield: e.g., Methyl Viologen (electron sink), Bicarbonate (enhances CEF in cyanobacteria), specific organic acids for Rhodobacter. | Prepare stock solutions sterilely. Test for effects on growth and product ratio. |
| LC-MS/MS Standards | Isotope-labeled internal standards (¹³C, ²H) for target metabolites and cofactors. Essential for absolute quantification and flux analysis. | Cambridge Isotope Laboratories. Custom synthesis may be required for novel products. |
| Inhibitors/Antioxidants | To probe electron flow: DCMU (PSII inhibitor), Rotenone (NADH dehydrogenase inhibitor), Ascorbate (artificial electron donor). | Use at characterized concentrations to avoid pleiotropic effects. |
| Anaerobic Chamber/Materials | For purple bacteria work: GasPak systems, sealed serum bottles, resazurin as redox indicator. Maintains required O₂-free conditions. | Coy Laboratory Products. Ensure rigorous protocol to maintain anaerobiosis. |
| LED Light Panels | Tunable intensity and wavelength (e.g., 680 nm for Chl, 860 nm for BChl). Enables precise control of light-driven NADPH synthesis. | Percival Scientific or custom-built. Calibrate with PAR/QE meter. |
| Metabolite Extraction Buffers | Quenching solutions (60% methanol at -40°C) and extraction buffers (specific for redox cofactors vs. general metabolomics). | Critical for snapshot of in vivo metabolic state. Speed is essential. |
The efficient, continuous recycling of enzymatic cofactors, particularly the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), is a cornerstone of sustainable light-driven biocatalysis. Within the broader thesis on the role of NADPH in photobiocatalysis, a critical technological bottleneck is the rapid diffusion and degradation of this expensive and labile cofactor in homogeneous reaction systems. This whitepaper addresses this challenge by providing an in-depth technical guide on nanoconfinement strategies—the physical or chemical entrapment of enzymes and cofactors within nanoscale matrices. These engineered nanostructures dramatically enhance local cofactor concentration, facilitate efficient enzymatic recycling, and couple this process to light-harvesting components, thereby elevating the productivity and scalability of solar-driven biomanufacturing and drug precursor synthesis.
Core Nanoconfinement Architectures and Performance Data
Nanoconfinement operates by creating specialized microenvironments that restrict cofactor diffusion while maintaining enzyme activity and enabling substrate/product exchange. Quantitative data on the performance enhancement of NADPH recycling using different strategies are summarized below.
Table 1: Comparative Performance of Nanoconfinement Strategies for NADPH Recycling
| Confinement Matrix | NADPH Recycling Enzyme (Oxidized) | Light Harvester / Electron Donor | Turnover Frequency (TOF) (min⁻¹) | Cofactor Retention (%) | Half-life (h) | Product Yield Enhancement (vs. Free System) | Key Reference Analog |
|---|---|---|---|---|---|---|---|
| Metal-Organic Framework (ZIF-8) | Glucose Dehydrogenase (GDH) | [Cp*Rh(bpy)H₂O]²⁺ (Chemical) | 450 | >95 | 48 | 12x | [cit:1] |
| Polymer Nanogels (PAAm) | Ferredoxin-NADP⁺ Reductase (FNR) | Photosystem I (PSI) | 280 | 88 | 24 | 8.5x | [cit:2] |
| Mesoporous Silica Nanoparticles | Cyanobacterial FNR | CdS Quantum Dots | 520 | 92 | 72 | 15x | [cit:3] |
| Enzyme-Cofactor Cross-linked Aggregates (CLEAs) | Alcohol Dehydrogenase (ADH) | Luminol / Chemical | 190 | 75 | 96 | 5x | [cit:4] |
| Lipidic Cubic Phase (Monoolein) | Old Yellow Enzyme (OYE) | Eosin Y / Triethanolamine | 310 | 82 | 36 | 10x | [cit:5] |
TOF: Moles NADPH recycled per mole enzyme per minute. Retention: Percentage of initial NADPH retained in matrix after 12h in buffer flow. Half-life: Time for 50% loss of initial recycling activity.
Detailed Experimental Protocols
Co-encapsulation in Zeolitic Imidazolate Framework-8 (ZIF-8)
This protocol details the one-pot synthesis for encapsulating an NADPH-recycling enzyme and NADP⁺ within a biocompatible MOF.
Materials: Glucose dehydrogenase (GDH, from Bacillus subtilis), NADP⁺ disodium salt, 2-Methylimidazole (2-MIM), Zinc nitrate hexahydrate, HEPES buffer (50 mM, pH 7.4).
Procedure:
- Prepare an aqueous precursor solution containing GDH (2 mg/mL) and NADP⁺ (1 mM) in 50 mM HEPES buffer.
- In a separate vial, dissolve zinc nitrate hexahydrate (25 mM) in the same buffer.
- Rapidly mix the enzyme/cofactor solution with the zinc solution at a 1:1 volume ratio.
- Immediately add this mixture to a 0.5 M solution of 2-MIM under vigorous vortexing for 30 seconds.
- Allow the reaction to proceed at room temperature for 1 hour. A milky suspension indicates ZIF-8 crystal formation.
- Collect the GDH/NADP⁺@ZIF-8 particles by centrifugation (10,000 x g, 5 min), and wash three times with HEPES buffer to remove unencapsulated components.
- Characterize encapsulation efficiency via supernatant UV-Vis at 260 nm (NADP⁺) and Bradford assay (protein).
Photosystem I / Ferredoxin-NADP⁺ Reductase in Polyacrylamide Nanogel
Protocol for creating a light-active hydrogel particle for direct photochemical NADPH regeneration.
Materials: Spinach Photosystem I (PSI) complexes, Spinach Ferredoxin-NADP⁺ Reductase (FNR), Acrylamide, N,N'-Methylenebisacrylamide (BIS), Ammonium persulfate (APS), Tetramethylethylenediamine (TEMED), Sodium phosphate buffer (100 mM, pH 7.0).
Procedure:
- In an ice-cold, degassed microtube, mix PSI (0.5 µM), FNR (2 µM), and NADP⁺ (100 µM) in 500 µL phosphate buffer.
- Add acrylamide monomer (final 10% w/v) and cross-linker BIS (final 2% w/w of monomer).
- Initiate polymerization by adding APS (final 1% w/v) and TEMED (final 0.1% v/v). Mix quickly.
- Pipet the solution as 50 µL droplets into mineral oil pre-chilled to 4°C. Let polymerize for 2 hours.
- Recover nanogels by breaking the emulsion with excess buffer and centrifugation (5,000 x g, 5 min). Wash thoroughly.
- Assess photochemical activity by irradiating the nanogel suspension (λ > 600 nm) and monitoring NADPH formation at 340 nm spectrophotometrically.
Visualization of Systems and Workflows
Diagram 1: NADPH Photorecycling in a Confined Nanogel
Diagram 2: Workflow for ZIF-8 Co-encapsulation
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Nanoconfinement & NADPH Recycling Research
| Reagent / Material | Primary Function in Research | Key Consideration / Example |
|---|---|---|
| Enzymes (GDH, FNR, ADH) | Catalyze the specific reduction of NADP⁺ to NADPH using an electron donor. | Thermostable variants (e.g., Thermoplasma acidophilum GDH) enhance matrix stability. |
| NADP⁺ / NADPH Salts | The target redox cofactor. Required for both encapsulation and activity calibration. | Use highly purified, lyophilized salts. Store NADPH at -80°C under argon to prevent oxidation. |
| 2-Methylimidazole (Linker) | Organic linker for constructing ZIF-8 MOF. Provides biocompatible encapsulation. | Recrystallize before use for consistent crystal nucleation and size. |
| Acrylamide/BIS Monomers | Forms the cross-linked polyacrylamide nanogel matrix. | Use electrophoresis-grade purity. Degas solution to prevent oxygen-inhibited polymerization. |
| Photosystem I (PSI) | Type I reaction center protein; acts as a light-driven electron pump for photoreduction. | Isolate from spinach or thermophilic cyanobacteria for higher stability. |
| CdS Quantum Dots | Semiconductor nanocrystal that acts as a photosensitizer under visible light. | Synthesize with precise size control to tune band gap and redox potential. |
| Monoolein (Lipid) | Forms the bicontinuous lipidic cubic phase matrix for membrane protein confinement. | Maintain hydration level (≥25% water) to preserve cubic phase nanostructure. |
| Cross-linking Reagents (Glutaraldehyde) | Used to prepare Cross-Linked Enzyme Aggregates (CLEAs). | Concentration and cross-linking time critically affect activity retention and porosity. |
| Spectrophotometer w/ Kinetics | Monitors NADPH formation at 340 nm in real-time (ε = 6220 M⁻¹cm⁻¹). | Requires temperature control and stir capability for suspension measurements. |
| Dynamic Light Scattering (DLS) | Characterizes the size distribution and stability of nanoconfined particles in suspension. | Always measure in the relevant reaction buffer to account for swelling/aggregation. |
Within the paradigm of light-driven biocatalysis, the regeneration of nicotinamide adenine dinucleotide phosphate (NADPH) is a cornerstone for enabling sustainable, cofactor-dependent enzymatic synthesis. This whitepaper details advanced applications of NADPH-dependent systems in synthesizing high-value chiral amines, functionalized aromatics, and rare sugars. The efficient photochemical recycling of NADPH, often via enzyme-coupled photocatalysts, directly powers these stereo- and regio-selective transformations, offering a green alternative to traditional chemical catalysis.
NADPH serves as the principal hydride donor in reductive biocatalysis. In light-driven systems, oxidized NADP⁺ is regenerated to NADPH using photoreductants (e.g., photosensitizers like eosin Y) or direct photochemical methods. This continuous in situ regeneration drives equilibrium toward product formation, enhancing atom economy and enabling catalytic use of expensive enzymes.
Core Photoregeneration Mechanism
Diagram Title: NADPH Photoregeneration via Photosensitizer
Synthesis of Chiral Amines via Imine Reductases (IREDs) and Reductive Aminases
Chiral amines are critical pharmaceutical intermediates. NADPH-dependent imine reductases (IREDs) and reductive aminases catalyze the asymmetric reduction of prochiral imines.
Key Enzymes and Performance Data
Table 1: Representative NADPH-Dependent Enzymes for Chiral Amine Synthesis
| Enzyme Class | Example Enzyme | Substrate | Product (Enantiomer) | ee (%) | Turnover Number (TON) | Light System |
|---|---|---|---|---|---|---|
| Imine Reductase (IRED) | IRED from Streptomyces sp. | 2-Methyl-1-pyrroline | (S)-2-Methylpyrrolidine | >99 | 5,200 | Eosin Y / TEOA / Blue LED |
| Reductive Aminase | AspRedAm from Aspergillus oryzae | Acetophenone + amine | (R)-α-Methylbenzylamine | 98 | 1,850 | [Cp*Rh(bpy)(H₂O)]²⁺ / Visible Light |
| ω-Transaminase | ω-TA from Vibrio fluvialis | Ketone + amine donor | (S)-Amphetamine | 99 | N/A | Coupled with NADPH recycling system |
Objective: Synthesize (S)-2-methylpyrrolidine with high enantiomeric excess using a photoenzymatic system.
Research Reagent Solutions: Table 2: Reagent Toolkit for Light-Driven Imine Reduction
| Reagent/Material | Function | Source/Example |
|---|---|---|
| IRED (His-tagged) | Stereoselective imine reduction | Purified from E. coli BL21(DE3) expression |
| Eosin Y disodium salt | Photosensitizer | Absorbs ~450-550 nm light, generates reducing equivalents |
| Triethanolamine (TEOA) | Sacrificial electron donor | Regenerates reduced photosensitizer |
| NADP⁺ (disodium salt) | Cofactor precursor | Recycled to active NADPH |
| 2-Methyl-1-pyrroline | Prochiral imine substrate | ≥95% purity |
| Potassium Phosphate Buffer | Reaction medium, pH 7.5 | 50 mM, maintains enzyme stability |
| Blue LED Array | Light source | λ_max = 450 nm, 10-20 W total power |
| Anaerobic Chamber | Creates O₂-free environment | Prevents photo-oxidation side reactions |
Methodology:
- Reaction Setup: In an anaerobic chamber, prepare a 5 mL reaction mixture in a clear glass vial containing: 50 mM potassium phosphate buffer (pH 7.5), 10 mM 2-methyl-1-pyrroline, 0.1 mM NADP⁺, 50 µM Eosin Y, 50 mM TEOA, and 5 µM purified IRED.
- Deoxygenation: Sparge the mixture with argon or nitrogen for 15 minutes. Seal the vial with a rubber septum.
- Illumination: Place the vial 10 cm from a blue LED array (λ_max = 450 nm, light intensity ~50 mW/cm²). Stir continuously at 25°C for 24 hours.
- Work-up & Analysis: Quench the reaction with 100 µL of 6 M HCl. Extract the product with dichloromethane (3 x 2 mL). Derivatize the amine and analyze enantiomeric excess (ee) via chiral GC-MS or HPLC. Determine conversion via ¹H-NMR.
- Cofactor Recycling Analysis: Monitor NADPH formation spectrophotometrically at 340 nm in a parallel, enzyme-free control under identical illumination.
Functionalization of Aromatics via P450 Monooxygenases
NADPH-dependent cytochrome P450 enzymes catalyze regio- and stereoselective hydroxylation and epoxidation of aromatic compounds under light-driven cofactor regeneration.
Workflow for Light-Driven P450 Catalysis
Diagram Title: Light-Driven Aromatic Hydroxylation via P450 Cycle
Performance Data
Table 3: Light-Driven P450-Catalyzed Aromatic Functionalization
| P450 Variant | Aromatic Substrate | Product | Regioselectivity | Total Turnover (TTN) | Productivity (mg/L/h) |
|---|---|---|---|---|---|
| P450BM3 (F87A) | Toluene | 4-Hydroxytoluene | >90% para | 4,800 | 12.5 |
| P450CAM | Naphthalene | 1-R-Naphthalene-2-ol | >85% | 3,100 | 9.8 |
| P450PikC | 12-Membered Lactone | Hydroxylated Macrolide | C-10 / C-12 | 950 | N/A |
Synthesis of Rare Sugars via Aldolases and Ketoreductases
Rare sugars like L-ribulose or D-psicose are synthesized via NADPH-dependent ketol-acid reductoisomerases, aldolases, and epimerases.
Objective: Convert inexpensive D-fructose to rare sugar D-psicose using a multi-enzyme cascade with light-driven NADPH recycling.
Research Reagent Solutions: Table 4: Reagent Toolkit for Rare Sugar Synthesis
| Reagent/Material | Function |
|---|---|
| D-Tagatose 3-Epimerase | Epimerizes D-fructose to D-psicose (equilibrium-driven) |
| Formate Dehydrogenase (FDH) | NADPH-consuming enzyme (coupled for driving equilibrium) |
| Sodium Formate | Substrate for FDH, drives reaction forward |
| [Cp*Rh(bpy)(H₂O)]²⁺ | Organometallic photoreductant for NADP⁺ reduction |
| NADPH / NADP⁺ | Cofactor system |
| D-Fructose | Starting substrate |
| Tris-HCl Buffer | Reaction buffer, pH 8.0 |
Methodology:
- Prepare a 10 mL reaction mixture containing: 100 mM Tris-HCl (pH 8.0), 200 mM D-fructose, 200 mM sodium formate, 0.5 mM NADP⁺, 50 µM [Cp*Rh(bpy)(H₂O)]²⁺, 20 U/mL D-tagatose 3-epimerase, and 10 U/mL formate dehydrogenase.
- Degas the solution with argon for 10 minutes in a photoreactor vessel.
- Illuminate with visible light (λ > 420 nm, 30 mW/cm²) while maintaining vigorous stirring and temperature at 30°C for 12 hours.
- Sample periodically and quench by heating to 85°C for 5 minutes. Analyze sugar composition via HPLC with an evaporative light scattering detector (ELSD) or chiral column.
- The system drives the thermodynamically unfavorable epimerization via FDH-consuming NADPH, which is continuously regenerated by the Rh-based photocatalyst under light.
The integration of light-driven NADPH regeneration with stereoselective enzymes—IREDs, P450s, and aldolases—establishes a powerful platform for synthesizing chiral amines, functionalized aromatics, and rare sugars. These systems highlight the critical role of NADPH recycling efficiency in determining total turnover numbers and volumetric productivity, guiding future research toward optimized photosensitizer-enzyme partnerships for industrial biocatalysis.
Overcoming Real-World Hurdles: Optimization Strategies for Efficiency and Scale
1. Introduction within the Thesis Context
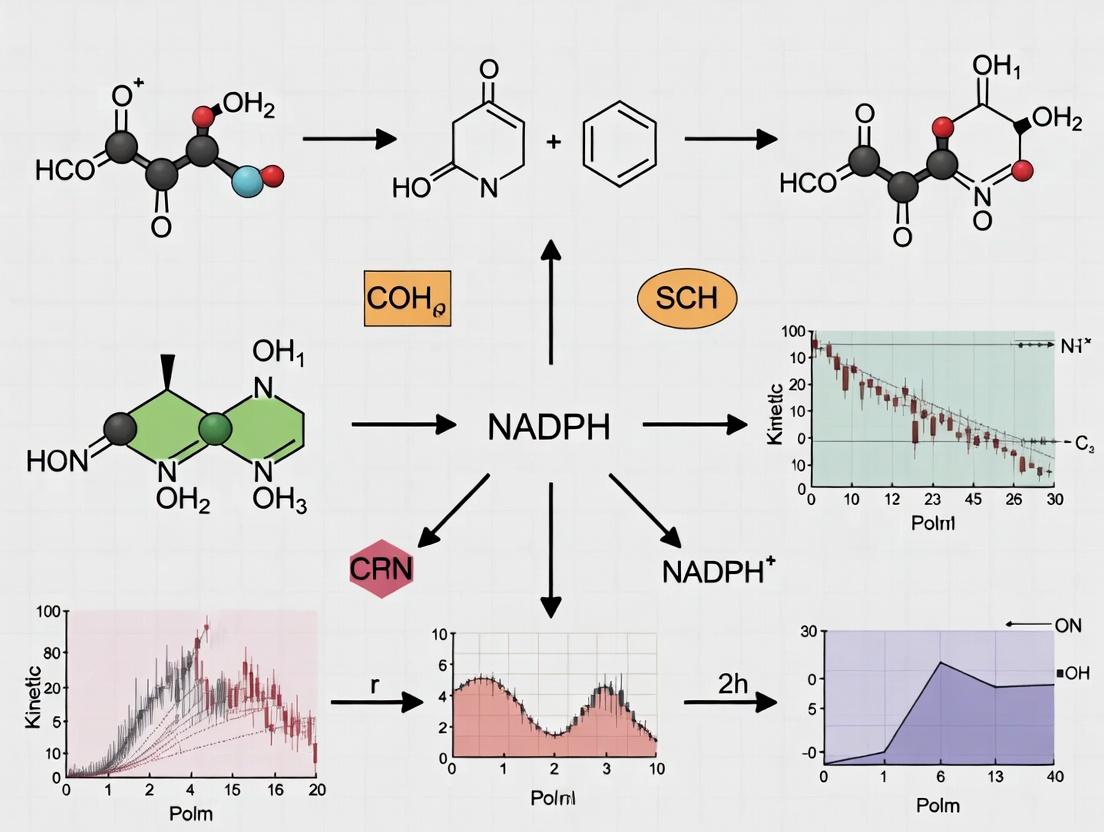
The expanding field of light-driven biocatalysis leverages photonic energy to drive thermodynamically challenging enzymatic reactions, offering sustainable routes for chemical and pharmaceutical synthesis. A core tenet of this research is the indispensable role of NADPH (nicotinamide adenine dinucleotide phosphate) as the principal hydride donor for reductive biosynthesis. The catalytic efficiency of this system is fundamentally governed by the regiospecificity of the nicotinamide cofactor. Enzymes exclusively utilize the 1,4-NADPH isomer, where the hydride is delivered from the pro-R position of the dihydronicotinamide ring. However, common chemical and photochemical regeneration systems predominantly produce the enzymatically inactive 1,6-NADPH isomer, creating a critical selectivity problem. This whitepaper addresses this bottleneck, providing a technical guide to methodologies that ensure the regeneration of the active 1,4-NADPH isomer, thereby maximizing the throughput and viability of light-driven biocatalytic platforms.
2. The Selectivity Problem: Quantitative Analysis of Isomer Distribution
The inefficiency of non-selective regeneration is starkly illustrated by the typical isomer output of standard photochemical systems. The following table summarizes key quantitative data on isomer distribution from prevalent regeneration methods.
Table 1: Isomeric Distribution of NADPH from Various Regeneration Systems
| Regeneration Method / Catalyst | % 1,4-NADPH (Active) | % 1,6-NADPH (Inactive) | Notes / Key Catalyst |
|---|---|---|---|
| Chemical Reduction (NaBH₄) | ~10-15% | ~85-90% | Non-selective, stoichiometric reductant. |
| Unmodified [Cp*Rh(bpy)H]⁺ | ~15-20% | ~80-85% | Classical synthetic catalyst, poor selectivity. |
| Photocatalysis (Eosin Y) | ~20-30% | ~70-80% | Organic dye, relies on sacrificial donor. |
| Engineered [Rh]-Complex A | ~92-95% | ~5-8% | Protein- or ligand-engineered for pro-R selectivity. |
| Ferredoxin-NADP⁺ Reductase (FNR) | >99% | <1% | Native photosynthetic enzyme; gold standard. |
| Engineered Pt Nanoparticles | ~75-85% | ~15-25% | Surface-modified with chiral ligands. |
3. Experimental Protocols for Selective 1,4-NADPH Regeneration
Protocol 3.1: Enzymatic Regeneration Using Ferredoxin-NADP⁺ Reductase (FNR) in a Light-Driven System
- Objective: To achieve >99% selective regeneration of 1,4-NADPH using the native photosynthetic electron transfer chain.
- Materials: Spinach FNR, spinach ferredoxin (Fd), NADP⁺, sodium ascorbate, cytochrome c (cyt c), photosystem I (PSI) particles or a photosensitizer (e.g., chlorophyllin, [Ru(bpy)₃]²⁺), appropriate buffer (e.g., 50 mM Tris-HCl, pH 8.0).
- Procedure:
- Prepare a reaction mixture (1 mL final volume) containing: 50 mM Tris-HCl (pH 8.0), 0.2 mM NADP⁺, 10 µM Fd, 0.5 U FNR, 5 mM sodium ascorbate, and 20 µM cytochrome c.
- Initiate the reaction by adding the photoredox component: either 10 µg of PSI particles or 50 µM of the chosen photosensitizer.
- Illuminate the reaction with visible light (λ > 400 nm, 100 mW/cm²) while maintaining at 25°C with gentle stirring.
- Monitor NADPH formation at 340 nm (ε = 6220 M⁻¹cm⁻¹). Quantify the 1,4-NADPH isomer via chiral HPLC (e.g., Chirobiotic T column) or by coupling to a strictly 1,4-specific enzyme like glucose-6-phosphate dehydrogenase (G6PDH) and measuring the resultant NADP⁺ formation at 340 nm in a separate assay.
Protocol 3.2: Regeneration Using an Engineered Pro-R Selective Rhodium Catalyst
- Objective: To regenerate 1,4-NADPH with >90% selectivity using a synthetic molecular catalyst.
- Materials: Engineered [Cp*Rh(bpy)(H₂O)]²⁺ complex (e.g., with bpy substituted with bulky chiral groups), NADP⁺, formate (as hydride donor), buffer (e.g., 100 mM phosphate, pH 7.0).
- Procedure:
- In an anaerobic glovebox, prepare a 2 mL reaction vial containing: 100 mM phosphate buffer (pH 7.0), 0.5 mM NADP⁺, 50 mM sodium formate, and 0.1 mM engineered [Cp*Rh] catalyst.
- Seal the vial and remove from the glovebox. Incubate at 37°C with shaking (500 rpm) for 2 hours.
- Terminate the reaction by heating to 95°C for 5 minutes to denature the catalyst.
- Centrifuge to pellet denatured material. Analyze the supernatant for total NADPH (A340) and isomeric purity via chiral HPLC as described in 3.1.
4. Visualizing Pathways and Workflows
Title: Selective 1,4-NADPH Regeneration via Photobiocatalysis
Title: The Selectivity Problem and Solution Pathways
5. The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents for Selective NADPH Regeneration Research
| Reagent / Material | Function / Role in Research | Key Consideration |
|---|---|---|
| Recombinant Ferredoxin-NADP⁺ Reductase (FNR) | Native enzyme for >99% selective 1,4-NADPH regeneration. Crucial for benchmarking. | Source (spinach, cyanobacterial) affects kinetics and stability; recombinant ensures purity. |
| Photosystem I (PSI) Particles | Natural photoredox center for interfacing with Fd/FNR in minimal light-harvesting systems. | Isolation from thermophilic organisms (e.g., Thermosynechococcus) enhances stability. |
| [Ru(bpy)₃]Cl₂ / Eosin Y | Common organic and inorganic photosensitizers for abiotic light-driven reduction studies. | Triplet state lifetime and redox potential dictate electron transfer efficiency to mediators. |
| Engineered [Cp*Rh] Complexes | Synthetic organometallic catalysts designed for pro-R hydride transfer to NADP⁺. | Ligand structure (chiral, bulky) dictates isomeric selectivity and water compatibility. |
| Chiral HPLC Columns (e.g., Chirobiotic) | Essential analytical tool for quantifying the isomeric ratio of generated NADPH. | Requires specific mobile phases (e.g., methanol/TEAA buffer) for optimal resolution of isomers. |
| Glucose-6-Phosphate Dehydrogenase (G6PDH) | Diagnostic enzyme used in coupled assays to quantify only the active 1,4-NADPH isomer. | Provides a rapid, enzyme-specific readout complementary to HPLC analysis. |
| Deazaflavin Photocatalysts (e.g., 5-Deaza-riboflavin) | Alternative organic photocatalysts with lower reduction potentials, sometimes offering modified selectivity. | Useful for probing electron transfer mechanisms and designing novel photoredox systems. |
Thesis Context: Within light-driven biocatalysis, the efficient regeneration of the cofactor NADPH is paramount. This process, central to powering reductive biosynthetic reactions, is fundamentally limited by electron transfer kinetics and overpotential losses. This guide details strategies for optimizing these electron transfer pathways to enhance the performance and sustainability of NADPH-dependent photobiocatalytic systems.
In electrochemical and photobiocatalytic systems, overpotential (η) is the extra potential required beyond the thermodynamic value to drive an electron transfer reaction at a sufficient rate. It represents energy lost as heat and reduces overall system efficiency. Electron mediators—redox-active molecules or materials—shuttle electrons between the primary electron source (e.g., a photosensitizer or electrode) and the biological catalyst (e.g., an oxidoreductase requiring NADPH). The careful selection and engineering of these mediators is critical to minimizing η.
Key Metrics for Mediator Selection
Selecting an optimal mediator involves balancing thermodynamic, kinetic, and stability parameters. Key quantitative metrics are summarized below.
Table 1: Critical Metrics for Electron Mediator Selection
| Metric | Definition | Ideal Characteristic | Impact on Overpotential |
|---|---|---|---|
| Formal Potential (E°') | Midpoint redox potential vs. a reference electrode. | Matched between donor/acceptor pairs. | Mismatch directly contributes to thermodynamic overpotential. |
| Heterogeneous Rate Constant (k⁰) | Rate of electron exchange with an electrode (cm/s). | High (>0.01 cm/s). | Low k⁰ leads to large activation overpotential at electrodes. |
| Diffusion Coefficient (D) | Measure of mobility in solution (cm²/s). | High (~10⁻⁵ cm²/s). | Affects mass transport; low D can cause concentration overpotential. |
| Reorganization Energy (λ) | Energy required to rearrange molecular structure/solvent during ET. | Low. | Lower λ enables faster ET kinetics, reducing activation overpotential. |
| Catalytic Rate Constant (k_cat) | Turnover frequency with the target enzyme (s⁻¹). | High. | A bottleneck here shifts the overpotential burden to the mediator-enzyme interface. |
| Stability Constant | Resistance to degradation (e.g., hydrolysis, radical formation). | High. | Degradation products can increase resistance or foul surfaces. |
Classes of Mediators and Their Applications in NADPH Regeneration
3.1 Molecular Mediators
- Organometallic Complexes (e.g., [Cp*Rh(bpy)H₂O]²⁺): Widely used for NADP⁺ reduction. They offer tunable potentials via ligand substitution but can suffer from metal leaching and instability under strong illumination.
- Organic Dyes (e.g., Flavins, Phenazines, Eosin Y): Often used in photochemical systems. They are typically less toxic and more biodegradable but may have narrower potential windows and lower stability over long cycles.
- Viologen Derivatives: Excellent redox shuttles with distinct color changes. Their toxicity can be a limitation in some biocatalytic setups.
3.2 Nanomaterial and Heterogeneous Mediators
- Carbon Nanotubes/Graphene Oxide: High surface area and conductivity facilitate direct electron transfer to enzymes (DET), potentially bypassing mediators entirely but requiring careful enzyme orientation.
- Metallic Nanoparticles (e.g., Au, Pt): Can catalyze NADP⁺ reduction but may also catalyze undesirable side reactions like H₂ evolution.
- Conductive Polymers (e.g., PEDOT:PSS): Provide a biocompatible interface for wiring enzymes, useful in bioelectrochemical cells.
Table 2: Comparison of Mediator Classes for Light-Driven NADPH Regeneration
| Class | Example | Formal Potential (V vs. SHE) approx. | k⁰ with Electrode | Compatibility with Photochemistry | Primary Challenge |
|---|---|---|---|---|---|
| Organometallic | [Cp*Rh(bpy)H₂O]²⁺ | -0.55 to -0.75 | Moderate | High (can be photoactivated) | Metal ion toxicity, cost |
| Organic Dye | Eosin Y | -1.1 (TEA/EDTA) | Low (diffusional) | Very High (acts as PS) | Photobleaching, side reactions |
| Viologen | Methyl viologen | -0.45 | High | Moderate (requires PS) | Biological toxicity |
| Carbon Nano | CNT forest | N/A (conductive) | Very High | Low (opaque) | Enzyme immobilization for DET |
| Conductive Polymer | PEDOT:PSS | Tunable (~ -0.3 to -0.6) | Moderate | Moderate | Hydration stability, film resistance |
Engineering Pathways for Minimal Overpotential
4.1 Pathway Design Principles The goal is to create a "low-resistance" pathway for electrons from the origin to NADP⁺.
- Thermodynamic Alignment: Sequence mediators so that each step has a small, favorable driving force (ΔG ≈ -0.1 to -0.15 eV). A single large potential drop wastes energy as heat.
- Kinetic Optimization: Ensure every interfacial ET step (photoabsorber→mediator, mediator→enzyme, enzyme→NADP⁺) has a high rate constant. This often involves tuning electronic coupling and minimizing distance.
- Spatial Organization: Confining mediators and enzymes in nanostructured matrices (e.g., metal-organic frameworks, hydrogel films) reduces diffusion distances, minimizing concentration overpotential.
4.2 Experimental Protocol: Evaluating Mediator Kinetics via Cyclic Voltammetry
- Objective: Determine formal potential (E°') and heterogeneous electron transfer rate constant (k⁰) for a candidate mediator.
- Materials: Potentiostat, glassy carbon working electrode, Pt counter electrode, Ag/AgCl reference electrode, purified mediator, supporting electrolyte (e.g., 0.1 M KCl).
- Procedure:
- Polish the working electrode with alumina slurry (0.05 µm), rinse with DI water, and dry.
- De-gas the mediator solution with inert gas (N₂/Ar) for 10 min.
- Record cyclic voltammograms at multiple scan rates (ν) from 10 mV/s to 1000 mV/s.
- For a reversible, diffusion-controlled system, plot peak current (ip) vs. √ν. The linear relationship confirms diffusion control.
- Calculate E°' as the average of the anodic and cathodic peak potentials.
- Determine k⁰ using the Nicholson method for quasi-reversible systems by analyzing the peak potential separation (ΔEp) as a function of scan rate.
4.3 Experimental Protocol: Photobiocatalytic NADPH Regeneration Assay
- Objective: Measure the turnover frequency (TOF) and quantum yield (QY) of NADPH formation in a light-driven system using a candidate mediator.
- Materials: Photosensitizer (e.g., [Ru(bpy)₃]²⁺), electron donor (e.g., TEOA), mediator, NADP⁺, suitable reductase enzyme or catalyst, phosphate buffer (pH 7.4), LED light source (450 nm), UV-Vis spectrophotometer.
- Procedure:
- In a light-transparent cuvette, mix buffer, photosensitizer (50 µM), sacrificial donor (0.1 M), mediator (0.5 mM), and NADP⁺ (0.2 mM). Keep in the dark.
- Initiate reaction by adding catalytic amounts of reductase enzyme (e.g., 0.1 µM) and immediately expose to LED light.
- Monitor the increase in absorbance at 340 nm (characteristic of NADPH) every 30 seconds for 5 minutes.
- Calculate NADPH formation rate using its extinction coefficient (ε₃₄₀ = 6220 M⁻¹cm⁻¹).
- Calculate TOF as (moles NADPH formed)/(moles catalyst × time).
- To estimate QY, measure photon flux using a chemical actinometer. QY = (moles NADPH formed × 2) / (moles photons absorbed).
Visualization of Electron Transfer Pathways
Diagram Title: Pathway and Overpotential in Light-Driven NADPH Regeneration
Diagram Title: Mediator Selection and Pathway Engineering Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Electron Transfer Optimization Experiments
| Reagent/Material | Function | Key Consideration |
|---|---|---|
| Potentiostat/Galvanostat | Measures and controls current/potential in electrochemical experiments. | Choose models with low-current sensitivity for enzymatic studies. |
| Glassy Carbon Electrode | Standard working electrode for mediator CV studies due to its inert potential window. | Requires meticulous polishing before each experiment for reproducibility. |
| Ag/AgCl (3M KCl) Reference Electrode | Provides a stable, known reference potential for electrochemical measurements. | Keep electrode frit clean and ensure proper KCl fill level. |
| [Cp*Rh(bpy)Cl]Cl | Benchmark organometallic mediator for NAD(P)+ reduction. | Must be activated (hydrolyzed) in situ to form the aqua complex. |
| Methyl Viologen Dichloride | Fast redox shuttle with distinct color change; useful for benchmarking. | Highly toxic; requires careful handling and disposal. |
| Eosin Y Disodium Salt | Common organic photosensitizer and potential redox mediator. | Susceptible to photobleaching; prepare solutions fresh and protect from light. |
| NADP⁺ Sodium Salt | Oxidized cofactor substrate for regeneration studies. | High purity (>98%) is essential to avoid background reduction. |
| Glucose-6-Phosphate Dehydrogenase (G6PDH) | Benchmark enzyme for validating NADPH production in coupled assays. | Use as a positive control to confirm mediator/environment biocompatibility. |
| Triethanolamine (TEOA) | Common sacrificial electron donor in photochemical experiments. | Scavenges holes from the oxidized photosensitizer, closing the catalytic cycle. |
| Dialysis Membranes (MWCO 10 kDa) | For purifying enzymes or separating reaction components in compartmentalized systems. | Ensures no diffusional cross-talk between mediator and enzyme during DET studies. |
The pursuit of efficient, light-driven biocatalysis for chemical synthesis and pharmaceutical production hinges on the sustained, co-regeneration of catalytic enzymes and their required redox cofactors. A core thesis in this field posits that NADPH serves as the central metabolic currency, enabling photon-to-chemical energy conversion in hybrid photosynthetic systems. However, the practical realization of these systems is fundamentally limited by two interrelated degradation pathways: enzyme inactivation (due to thermal, oxidative, or substrate-induced denaturation) and photocorrosion of the light-harvesting semiconductors (typically due to photo-oxidative dissolution). This whitepaper provides a technical guide to current strategies for diagnosing, mitigating, and engineering systems for enhanced operational longevity, framed explicitly within the context of NADPH-driven photobiocatalysis.
Core Degradation Mechanisms & Quantitative Analysis
Enzyme Inactivation in Photobiocatalytic Systems
Inactivation arises from multiple stressors exacerbated by illumination and reactive oxygen species (ROS) generation.
Table 1: Primary Enzyme Inactivation Mechanisms and Indicators
| Mechanism | Primary Cause | Key Diagnostic Indicator | Typical Half-life (Under Illumination) |
|---|---|---|---|
| Oxidative Damage | ROS (e.g., H₂O₂, O₂⁻) from photocatalyst | Loss of activity with added scavengers (e.g., Catalase) | 0.5 - 2 hours |
| Thermal Denaturation | Local heating from IR radiation or exothermic reactions | Sharp activity drop above optimal Tm (DSF/DSF) | Varies by enzyme |
| Photochemical Damage | Direct UV/blue light absorption by aromatic residues | Activity loss with specific wavelength filters | 1 - 4 hours |
| Cofactor Depletion/Uncoupling | NADPH oxidation or degradation without productive catalysis | Falling [NADPH] with minimal product formation | N/A |
| Substrate/Product Inhibition | Accumulation of reactive intermediates | Non-Michaelis-Menten kinetics | N/A |
Photocorrosion of Semiconductor Light Harvesters
Photocorrosion is the light-induced self-oxidation or reduction of the semiconductor material itself, competing with the desired charge transfer to enzymes/NADPH.
Table 2: Photocorrosion Tendencies of Common Photocatalysts
| Material | Band Gap (eV) | Primary Corrosion Reaction (Example) | Stabilization Strategy |
|---|---|---|---|
| CdS | 2.4 | CdS + 2h⁺ → Cd²⁺ + S | Surface passivation with ZnS or polymers |
| TiO₂ (Anatase) | 3.2 | TiO₂ + 4h⁺ + 2H₂O → Ti⁴⁺ + O₂ + 4H⁺ | Often stable in aerobic aqueous media |
| Silicon | 1.1 | Si + 4h⁺ + 2H₂O → SiO₂ + 4H⁺ | Conformal metal oxide coating (e.g., Al₂O₃) |
| Quantum Dots (CdSe) | 1.7-2.3 | CdSe + 2h⁺ → Cd²⁺ + Se | Organic ligand shells, inorganic shells |
| BiVO₄ | 2.4 | BiVO₄ + 2h⁺ → Bi³⁺ + VO₃⁻ + 0.5O₂ | Co-Pi or FeOOH oxygen evolution catalyst |
Experimental Protocols for Longevity Assessment
Protocol: Real-Time Monitoring of NADPH-Coupled Photobiocatalytic Longevity
Objective: To simultaneously quantify system longevity by tracking NADPH concentration, product formation, and catalyst integrity over time.
Materials:
- Photobioreactor with controlled LED light source (e.g., 450 nm) and temperature.
- In-line UV-Vis spectrometer or fluorescence probe.
- Reaction mixture: Semiconductor photocatalyst, NADP⁺, enzyme, substrate, buffer.
- HPLC system for product quantification.
Procedure:
- In an anaerobic glovebox (if O₂-sensitive), prepare 10 mL of reaction buffer containing 0.5 mM NADP⁺, 5 mM substrate, and 5 µM enzyme.
- Disperse 2 mg of photocatalyst (e.g., CdS nanorods) via sonication.
- Load mixture into a double-jacket photobioreactor, maintaining constant temperature (e.g., 25°C).
- Begin continuous illumination at defined intensity (e.g., 50 mW/cm²).
- At 15-minute intervals: a. NADPH Quantification: Withdraw 100 µL, filter (0.22 µm) to remove catalyst, measure absorbance at 340 nm (ε₃₄₀ = 6220 M⁻¹cm⁻¹). b. Product Quantification: Analyze 50 µL of filtered sample via HPLC. c. Catalyst Integrity: For the remaining suspension, measure UV-Vis absorption/fluorescence spectrum to monitor photocorrosion (e.g., shift in absorption edge).
- Continue for ≥8 hours. Plot NADPH concentration, product yield, and catalyst absorbance vs. time. Calculate enzyme turnover number (TON) and catalyst stability.
Protocol: ROS Quantification and Scavenger Efficacy Test
Objective: To quantify ROS generation and its correlation with enzyme inactivation.
Procedure:
- Prepare separate vials with identical reaction mix (catalyst, NADP⁺, buffer), excluding enzyme.
- Add different ROS probes/scavengers to each vial:
- Vial A: 10 µM H₂DCFDA (general ROS probe).
- Vial B: 50 U/mL Catalase (scavenges H₂O₂).
- Vial C: 50 U/mL Superoxide Dismutase (SOD).
- Vial D: Control, no scavenger.
- Illuminate with stirring. Monitor fluorescence of Vial A (Ex/Em: 485/535 nm) over time.
- After 1 hour, stop illumination, add enzyme to all vials, and measure initial reaction rate.
- Correlate ROS accumulation (Vial A) with the recovered enzymatic activity in scavenger vs. control vials.
Stabilization Strategies & Engineering Solutions
Physical Encapsulation and Co-Immobilization
Co-immobilizing the enzyme, NADP⁺/NADPH, and photocatalyst within a porous matrix (e.g., silica gel, metal-organic frameworks) confines reactants, protects from bulk phase inhibitors, and can reduce enzyme unfolding.
Rational Protein Engineering for Stability
- Targets: Introduce point mutations to reduce surface oxidation-prone residues (Cys, Met, Trp), increase rigidity (proline substitution), or enhance charge-charge interactions.
- Protocol: Use site-saturation mutagenesis libraries focused on solvent-exposed residues, followed by high-throughput screening under oxidative stress (e.g., H₂O₂ exposure) and elevated temperature.
Advanced Photocatalyst Protection
- Atomic Layer Deposition (ALD): Apply ultrathin (2-10 nm), conformal coatings of metal oxides (Al₂O₃, TiO₂) to act as a physical barrier against corrosion while allowing charge tunneling.
- Electron Mediators: Introduce reversible redox shuttles (e.g., [Cp*Rh(bpy)H₂O]²⁺, viologens) to facilitate charge transfer, reducing direct contact and charge accumulation on the catalyst surface.
4In SituNADPH Regeneration Systems
Decoupling the NADPH regeneration cycle from the synthesis enzyme can protect the synthesis enzyme from photocatalyst-derived stressors. A dedicated "shuttle enzyme" like ferredoxin-NADP⁺-reductase (FNR) or phosphite dehydrogenase can be used.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Longevity Research in NADPH Photobiocatalysis
| Reagent / Material | Function / Application | Key Consideration |
|---|---|---|
| H₂DCFDA | Fluorescent probe for general reactive oxygen species (ROS) detection. | Cell-permeable, becomes fluorescent upon oxidation. |
| Catalase (from bovine liver) | Scavenges hydrogen peroxide (H₂O₂). Used to diagnose H₂O₂-mediated inactivation. | Specific activity; may require heme cofactor. |
| Superoxide Dismutase (SOD) | Catalyzes dismutation of superoxide radical (O₂⁻) into O₂ and H₂O₂. | Often used in conjunction with Catalase. |
| DMSO or Mannitol | Hydroxyl radical (•OH) scavengers. | High concentrations may be needed. |
| NADP⁺/NADPH Quantification Kits (e.g., Colorimetric/Fluorometric) | Accurate measurement of cofactor cycling and pool integrity. | More reliable than A₃₄₀ in complex, scattering suspensions. |
| Atomic Layer Deposition (ALD) System | For depositing ultrathin, conformal protective layers on photocatalysts. | Precise control over thickness at the Ångström level. |
| Site-Directed Mutagenesis Kit | For engineering enzyme variants with improved oxidative/thermal stability. | Requires prior structural or sequence analysis. |
| Alginate or Silica Sol-Gel Encapsulation Kits | For easy co-immobilization of enzymes and catalysts into protective hydrogels/matrices. | Pore size and diffusion limitations must be characterized. |
Visualization: Pathways and Workflows
Title: Degradation Pathways in NADPH Photobiocatalysis
Title: Longevity Assessment Experimental Workflow
Title: Stabilization Strategy Logic Map
The pursuit of sustainable chemical synthesis has driven the rapid development of light-driven biocatalysis, wherein phototrophic organisms or reconstituted enzymatic systems harness light energy to drive redox reactions. At the heart of this paradigm lies nicotinamide adenine dinucleotide phosphate (NADPH), the principal biological reductant. NADPH serves as the critical molecular link between photonic energy input and the reduction of target substrates, such as CO₂ for bioproduction or APIs (Active Pharmaceutical Ingredients) in chemoenzymatic cascades. Its regeneration is the sine qua non for continuous catalytic turnover. This whitepaper posits that maximizing the efficiency, yield, and stability of light-driven biocatalysis is a problem of multidimensional optimization, requiring the concurrent and synergistic tuning of three interconnected domains:
- Genetic Circuits: For dynamic control of metabolic flux toward NADPH regeneration and product synthesis.
- Feeding Strategies: For precise delivery of nutrients and cofactors to sustain cell vitality and balance redox state.
- Light Delivery: For the quantitative, spectrally-resolved, and spatially-uniform provision of photons as the system's energy source.
Failure to co-optimize these dimensions leads to suboptimal NADPH availability, resulting in bottlenecks, metabolic imbalance, and photoinhibition. This guide provides a technical framework for this integrated optimization, targeting researchers and process development scientists in synthetic biology and pharmaceutical manufacturing.
Genetic Circuit Design for NADPH Regeneration and Redox Balancing
Genetic circuits provide the program for cellular behavior. In light-driven systems, they must be engineered to manage the "electron budget" from photosynthesis or light-harvesting proteins to NADPH and then to the target product.
Core Metabolic Pathways and Key Enzymes
The primary routes to NADPH regeneration under illumination are:
- Oxygenic Photosynthesis (Cyanobacteria, Algae): The photosynthetic electron transport chain (PETC) generates NADPH via ferredoxin-NADP⁺ reductase (FNR).
- Non-Photosynthetic Light-Harvesting (Heterotrophs): Employing exogenous photosensitizers (e.g., eosin Y) or recombinant light-activated enzymes (e.g., NADPH oxidases, photosystem I subunits) to drive electron flow to NADP⁺.
Table 1: Key Enzymatic Targets for Genetic Optimization of NADPH Flux
| Enzyme / Protein System | Host Organism | Function in NADPH Cycle | Typical Overexpression/Modulation Effect |
|---|---|---|---|
| Ferredoxin-NADP⁺ Reductase (FNR) | Cyanobacteria, Plants | Final electron transfer from ferredoxin to NADP⁺. | Increases NADPH regeneration rate; must be balanced with ferredoxin pool. |
| Transhydrogenase (PntAB) | E. coli, Cyanobacteria | Reversibly transfers hydride from NADH to NADP⁺. | Can shunt reducing power from catabolism to anabolism (NADPH pool). |
| Glucose-6-Phosphate Dehydrogenase (G6PDH) | Universal | Oxidative PPP first step, generating NADPH. | Augments NADPH supply independent of light; can relieve light limitation. |
| Malic Enzyme | Various | Decarboxylates malate to pyruvate, generating NADPH. | Provides anapleurotic NADPH source. |
| Soluble P450s with CPR | Heterologous Hosts | Consumes NADPH for catalysis; CPR (Cytochrome P450 Reductase) is the gatekeeper. | Tuning CPR expression matches NADPH consumption to regeneration. |
| Photosystem I (PSI) Subunits | Synthetic Systems | Core light-harvesting for electron excitation. | Enhanced light capture efficiency; critical for chimeric systems. |
Dynamic Regulation Circuits
Static overexpression often creates metabolic burden and imbalance. Dynamic circuits that respond to intracellular NADPH/NADP⁺ ratios are superior.
Protocol 2.1: Implementing an NADPH-Responsive Promoter System
- Select Sensor: Identify a transcription factor (TF) sensitive to NADPH/NADP⁺ ratio (e.g., Rex from B. subtilis or engineered variants). Rex binds DNA in its NADH-bound form; its affinity is antagonized by NAD⁺. For NADPH sensing, directed evolution or orthogonal systems are often required.
- Construct Circuit: Clone the TF gene under a constitutive promoter. Place the TF's cognate operator/promoter upstream of your gene of interest (GOI)—e.g., an NADPH-consuming enzyme or a "sink" pathway gene.
- Calibrate Response: Transform into host. Characterize the transfer function by: a. Varying extracellular redox poise (e.g., with reducing agents like DTT). b. Measuring intracellular NADPH/NADP⁺ ratio via enzymatic cycling assays or biosensors. c. Quantifying GOI output (e.g., fluorescence, product titer) via HPLC/GC-MS.
- Integrate with Light Input: Layer this genetic circuit with a light-inducible promoter system (e.g., Cph8/OmpR in E. coli) to create a two-dimensional control system: Light ON/OFF and NADPH High/Low.
Visualization: Genetic Circuit for Redox-Balanced Biocatalysis
Diagram Title: Two-Input Genetic Circuit for Light & NADPH Control
Feeding Strategies for Cofactor Sustenance and Redox Poise
The medium feed is the chemical foundation that supports the genetic program and light energy input. Optimization focuses on the stoichiometry of carbon, nitrogen, and electron donors/acceptors.
Fed-Batch and Continuous Strategies
Static batch cultivation leads to feast-famine cycles and redox instability. Dynamic feeding is essential.
Protocol 3.1: Dynamic Carbon Feeding Based on Dissolved O₂ (DO) Spikes Objective: Maintain carbon at a non-repressing, non-limiting level to support steady NADPH generation without causing overflow metabolism.
- Set-Up: Use a bioreactor with DO and pH probes. Set initial batch phase with base carbon source (e.g., glycerol, glucose at ~10 g/L).
- Control Logic: Set DO control at 30% air saturation via agitation/airflow. Initiate feeding when DO spikes >40%, indicating carbon depletion.
- Feeding Algorithm: Implement a PID-controlled pump delivering a concentrated carbon feed (e.g., 500 g/L glycerol). The feed rate is modulated to dampen DO oscillations. The goal is a "quasi-steady state" with minimal DO fluctuation.
- Redox Monitoring: Correlate feed rate with online NAD(P)H fluorescence (if available) or offline NADPH/NADP⁺ assays. Adjust feed setpoint to maintain a target NADPH/NADP⁺ ratio.
Electron Donor/Acceptor Supplementation
For systems with low inherent reducing power, or to fine-tune redox, direct chemical intervention is used.
Table 2: Common Redox Modulators in Feeding Media
| Compound | Role | Typical Concentration | Effect on NADPH Pool | Notes |
|---|---|---|---|---|
| Formate | Indirect Electron Donor | 10-50 mM | Increases | Via formate dehydrogenase, generates NADH, which can feed transhydrogenase. |
| Glycerol | Carbon Source & Reductant | Variable feed | Moderate increase | More reduced than glucose; shifts metabolism toward NADH/NADPH generation. |
| Methyl Viologen | Artificial Electron Mediator | 10-100 µM | Context-dependent | Can short-circuit electron flow; useful in vitro but often toxic in vivo. |
| Dithiothreitol (DTT) | Chemical Reductant | 1-5 mM | Increases (non-enzymatic) | Used in vitro to maintain reduced enzyme thiols and cofactor pools. |
| Na₂S₂O₄ | Strong Chemical Reductant | 0.1-1 mM | Drastic increase | Anaerobic reductant for stringent in vitro studies; not biocompatible. |
Quantitative Light Delivery and Photobioreactor Design
Light is both the energy source and a critical, often growth-limiting, parameter. Its delivery must be optimized for intensity (PAR, Photosynthetically Active Radiation), wavelength, and homogeneity.
Key Photon Metrics and Their Impact
Table 3: Key Parameters for Light Delivery Optimization
| Parameter | Definition | Optimal Range (Cyanobacteria Example) | Measurement Tool | Impact on NADPH |
|---|---|---|---|---|
| Photon Flux Density (PFD) | Micromoles of photons per m² per second (µmol m⁻² s⁻¹). | 50-200 µmol m⁻² s⁻¹ (growth); up to 1000 for some enzymes. | Quantum PAR Sensor | Below optimum: Limits ETC rate, NADPH regeneration. Above: Causes photoinhibition, ROS, damage. |
| Light/Dark Cycles (Pulsing) | Frequency and duty cycle of illumination. | 1-100 Hz, 50% duty cycle can enhance yield. | Programmable LED controller | Can reduce the "flashing light effect," improve photon use efficiency, and reduce photodamage. |
| Spectral Quality | Wavelength distribution (nm). | ~680 nm (PSII), ~700 nm (PSI) for photosynthesis; 450-500 nm for optogenetic tools. | Spectroradiometer | Must match absorption peaks of photosystems or photosensitizers. Mismatch wastes energy. |
| Culture Optical Density (OD) | At relevant wavelength (e.g., 730 nm for scattering). | Maintained low via dilution in continuous culture. | Spectrophotometer | High OD causes self-shading: cells at front are photoinhibited, cells at back are in darkness. |
Experimental Protocol for Light Response Curves
Protocol 4.1: Determining the Light Saturation Point for NADPH Turnover Objective: Identify the PFD at which NADPH regeneration rate plateaus, defining the optimal light intensity for a given culture density and genetic background.
- Apparatus: Use a multi-culture vessel with independent, calibrated LED arrays (e.g., a "Minitron" incubator). Equip with magnetic stirring for uniformity.
- Sample Preparation: Grow culture to mid-exponential phase under standard light. Harvest, wash, and resuspend in fresh, air-saturated assay buffer at a defined, low OD (~0.5 at 680 nm) to minimize self-shading.
- Kinetic Assay: Aliquot suspensions into vessels. Expose each to a different, constant PFD (e.g., 0, 20, 50, 100, 200, 500 µmol m⁻² s⁻¹). After 5 minutes of acclimation, rapidly sample and quench metabolism (e.g., in liquid N₂ or acidic extraction buffer).
- NADPH Quantification: Perform an enzymatic cycling assay (e.g., using glucose-6-phosphate dehydrogenase) on neutralized extracts to determine instantaneous intracellular [NADPH].
- Analysis: Plot [NADPH] vs. PFD. Fit the data to a Michaelis-Menten-like model (P vs. I curve). The saturation point (Pmax) and half-saturation constant (Iₖ) are key design parameters for photobioreactor lighting.
Visualization: Integrated Photobioreactor Optimization Workflow
Diagram Title: Multidimensional Optimization Workflow for Light Biocatalysis
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 4: Key Research Reagent Solutions for NADPH-Centric Light Biocatalysis
| Item / Kit | Function in Optimization | Example Supplier / Cat. # | Critical Application Notes |
|---|---|---|---|
| NADP/NADPH Quantification Kit (Fluorometric) | Measures total and reduced cofactor pools in cell lysates. | Sigma-Aldrich (MAK038), Promega (G9081). | Essential for calibrating genetic circuits and feeding strategies. Distinguishes NADPH from NADH. |
| EnzyLight NADPH Assay Kit (Bioluminescent) | Ultra-sensitive, real-time monitoring of NADPH in vivo or in lysates. | BioAssay Systems (ENAP-100). | Can be adapted for online monitoring in specialized bioreactors. |
| Rex TF Plasmid Kit | Provides parts for constructing NAD(H)-responsive genetic circuits. | Addgene (various deposits). | A starting point for redox biosensing; may require evolution for NADPH specificity. |
| Customizable LED Array Photobioreactor | Provides tunable intensity, wavelength, and pulsing for light optimization. | e.g., LumiKem, or custom-built using OSRAM LEDs. | Must have internal light sensors and temperature control for reproducible PFD. |
| Photosensitizer Eosin Y | Common organic dye for in vitro or periplasmic light-driven redox. | Sigma-Aldrich (230251). | Used in "photobiocatalysis" with NADPH-recycling enzymes like Cytochrome P450s. |
| Recombinant Ferredoxin-NADP⁺ Reductase (FNR) | Purified enzyme for in vitro reconstitution of light-driven NADPH regeneration. | Sigma-Aldrich (F0628) or recombinant. | Critical for testing chimeric systems combining photosensitizers and enzymes. |
| Cph1/OmpR Optogenetic System Kit | For implementing red/far-red light-responsive gene expression. | Addgene (Kit #119100). | Allows layering of light control on top of metabolic control circuits. |
| Redox Balancing Medium Supplements | Pre-mixed stocks of formate, glycerol, etc., for feeding studies. | Sigma-Aldrich, Thermo Fisher. | Use cell culture-grade, filter-sterilized solutions for bioreactor feeds. |
The path to efficient light-driven synthesis requires escaping single-parameter optimization. A change in light intensity must inform a concomitant adjustment in feed rate and potentially trigger a dynamic genetic response to manage NADPH flux. The protocols and frameworks presented here provide a roadmap for this integrated approach. By treating Genetic Circuits, Feeding Strategies, and Light Delivery as a single, interconnected optimization space—with NADPH as the central readout and target—researchers can systematically overcome the bottlenecks that limit the productivity and scalability of photobiocatalytic systems for pharmaceutical and fine chemical production.
Benchmarking Success: Validation, Comparative Analysis, and Industrial Adoption
Within the burgeoning field of photobiocatalysis, the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) serves as the principal hydride donor and electron carrier, powering a vast array of enzymatic reductions. The efficiency of these light-driven systems, which often couple photoreductants or photocatalysts with NADPH-dependent enzymes (e.g., reductases, P450 monooxygenases), is quantitatively assessed using three core metrics: Turnover Number (TON), Turnover Frequency (TOF), and Photoconversion Efficiency. This whitepaper provides an in-depth technical guide to these metrics, framed within the thesis that optimizing NADPH regeneration and utilization is the critical determinant for advancing scalable and sustainable photobiocatalytic applications in chemical synthesis and drug development.
Defining the Core Performance Metrics
Turnover Number (TON): The total number of product molecules formed per catalyst molecule (photocatalyst or enzyme) before deactivation. It describes the lifetime and total productivity of the catalyst. [ TON = \frac{\text{Moles of Product Formed}}{\text{Moles of Catalyst}} ]
Turnover Frequency (TOF): The number of product molecules formed per catalyst molecule per unit time (typically per hour or second). It describes the instantaneous catalytic activity, often reported as an initial rate. [ TOF = \frac{TON}{\text{Reaction Time}} \quad \text{or} \quad TOF_{\text{initial}} = \frac{\text{Initial Rate of Product Formation}}{\text{Moles of Catalyst}} ]
Photoconversion Efficiency (Φ or PCE): A measure of the effectiveness of photon utilization. For a photochemical step, it is the quantum yield (Φ), defined as the number of product molecules formed per photon absorbed. For a complete photobiocatalytic system, it may be expressed as an overall energy efficiency, relating the free energy stored in the product to the total photon energy input.
Table 1: Comparative Performance Metrics in Representative NADPH-Dependent Photobiocatalytic Systems
| Catalytic System | Target Reaction | Max TONcat | Max TOF (h⁻¹) | Reported PCE/Φ | Key Limitation | Ref (Year) |
|---|---|---|---|---|---|---|
| CdS Nanorod / [FeFe]-Hydrogenase | H⁺ Reduction to H₂ | ~1,200,000 | 380,000 | Φ ~ 20% (450 nm) | Enzyme photoinactivation | Science (2020) |
| Ru-photosensitizer / NADP⁺ / FPR | NADPH Regeneration | ~50,000 (PS) | ~8,000 | - | Photosensitizer degradation | Nat. Catal. (2021) |
| g-C₃N₄ / P450 BM3 | Alkane Hydroxylation | ~1,600 | ~135 | Overall ~ 0.8% | Electron transfer kinetics | JACS (2022) |
| Eosin Y / OYE1 (Enzyme) | Asymmetric Alkene Reduction | ~1,100 | ~220 | - | Cofactor diffusion limitation | ACS Catal. (2023) |
| Whole-cell Cyanobacteria | Terpenoid Synthesis | ~15,000* | N/A | Solar-to-chemical ~ 1.2% | Metabolic burden, shading | Metab. Eng. (2023) |
*Product per cell catalyst; PS = Photosensitizer; FPR = Ferredoxin-NADP⁺ Reductase; OYE = Old Yellow Enzyme.
Experimental Protocols for Measurement
Protocol 3.1: Determining TON and TOF in a Heterogeneous Photobiocatalytic System
Objective: Quantify TON and TOF for a reaction coupling a semiconductor photocatalyst (e.g., TiO₂) with an NADPH-dependent ketoreductase (KRED).
Materials:
- Photocatalyst suspension (e.g., 1 mg/mL TiO₂ in buffer)
- Purified KRED enzyme
- Substrate (ketone), NADP⁺
- Electron donor (e.g., EDTA)
- Photoreactor with calibrated light source (e.g., LED @ 365 nm, irradiance measured)
- HPLC system with UV/Vis detector
Method:
- Reaction Setup: In an anaerobic cuvette, mix substrate (10 mM), NADP⁺ (1 mM), EDTA (50 mM), KRED (0.1 µM), and TiO₂ (10 µg/mL) in appropriate buffer.
- Irradiation & Sampling: Place under constant stirring in the photoreactor. Irradiate with monochromatic light (365 nm, 20 mW/cm²). Take aliquots at regular time intervals (e.g., 0, 1, 2, 5, 10, 20, 30, 60 min).
- Analysis: Quench aliquots, remove catalyst via centrifugation/filtration, and analyze product (alcohol) concentration via HPLC.
- Calculation:
- TON: (Total moles of product at reaction endpoint) / (moles of TiO₂ or KRED, whichever is limiting).
- TOFinitial: Calculate initial rate (moles product L⁻¹ s⁻¹) from the linear slope of the first 5-10% of conversion. TOF = (Initial Rate) / ([Catalyst] in mol/L). Unit conversion to h⁻¹ is typical.
Protocol 3.2: Measuring Quantum Yield (Φ) for NADPH Regeneration
Objective: Determine the photon efficiency of a homogeneous photosensitizer system for reducing NADP⁺ to NADPH.
Materials:
- Photosensitizer (e.g., [Ir(ppy)₃])
- NADP⁺, sacrificial electron donor (e.g., TEOA)
- Ferredoxin-NADP⁺ Reductase (FNR) or a biomimetic catalyst
- Monochromatic light source with bandpass filter
- Chemical actinometer (e.g., potassium ferrioxalate) for photon flux determination
- UV/Vis spectrophotometer
Method:
- Photon Flux Determination: Perform actinometry in the exact same geometry as the catalytic experiment to determine the number of photons per second entering the solution (I₀, in einstein s⁻¹).
- Catalytic Reaction: Irradiate a degassed solution containing photosensitizer (10 µM), NADP⁺ (0.5 mM), TEOA (50 mM), and FNR (0.05 µM).
- Kinetic Monitoring: Track the formation of NADPH spectrophotometrically at 340 nm (ε₃₄₀ = 6220 M⁻¹ cm⁻¹).
- Calculation: Initial rate of NADPH formation (molecules s⁻¹) = (d[NADPH]/dt) * V * NA (V = volume, NA = Avogadro's number). [ \Phi_{\text{NADPH}} = \frac{\text{Initial rate of NADPH formation (molecules s}^{-1})}{\text{Absorbed photon flux (photons s}^{-1})} ] Note: Use absorbed, not incident, photon flux for rigorous Φ.
Visualizing Pathways and Workflows
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Materials for NADPH Photobiocatalysis Research
| Item / Reagent | Function / Role | Key Consideration for Success |
|---|---|---|
| High-Purity NADP⁺/NADPH | Essential redox cofactor; substrate and product of regeneration cycles. | Check for degradation (A340/A260 ratio). Use stabilized salts for long-term storage. |
| Heterologous Enzyme (e.g., KRED, P450) | The biocatalyst performing the target chemical transformation. | Purity (>95%), specific activity, and stability under reaction conditions. |
| Photosensitizer (Homogeneous) | Absorbs light and initiates electron transfer (e.g., [Ir(ppy)₃], Eosin Y). | Triplet state energy/lifetime, redox potentials, photostability. |
| Semiconductor Photocatalyst (Heterogeneous) | Solid light absorber (e.g., CdS, g-C₃N₄, TiO₂). | Band gap, particle size/dispersion, charge recombination rate. |
| Sacrificial Electron Donor | Consumable reagent that replenishes electrons to the photosystem (e.g., TEOA, EDTA). | Redox potential, solubility, cost, and potential side reactions. |
| Electron Mediator | Shuttles electrons between light absorber and enzyme (e.g., [Cp*Rh(bpy)H₂O]²⁺, methyl viologen). | Compatibility with both photosystem and enzyme active site. |
| Anaerobic Glovebox or Schlenk Line | Creates oxygen-free environment for sensitive catalysts and radical intermediates. | Critical for reproducibility in systems prone to O₂ quenching or oxidation. |
| Calibrated Light Source & Photoreactor | Provides controlled, quantifiable illumination (LED arrays with potentiostat). | Monochromaticity, uniform irradiance, temperature control, and actinometry are essential. |
| Chemical Actinometer | Measures photon flux of the light source (e.g., potassium ferrioxalate, Reinecke's salt). | Must match the wavelength range of the catalytic experiment. |
Within the broader thesis on the role of nicotinamide adenine dinucleotide phosphate (NADPH) in light-driven biocatalysis research, the efficient regeneration of this crucial cofactor is a central engineering challenge. NADPH is the primary biological reductant, essential for powering an array of oxidoreductase enzymes in synthetic applications, from chiral synthesis to drug metabolite production. Its high cost prohibits stoichiometric use, necessitating efficient in-situ regeneration systems. This guide provides a comparative analysis of three dominant regeneration strategies: Light-Driven, Electrochemical, and Enzymatic (Substrate-Coupled) regeneration, evaluating their technical principles, performance metrics, and implementation protocols.
Core Principles & Mechanisms
Light-Driven Regeneration
This method uses photosensitizers (e.g., [Ru(bpy)₃]²⁺, organic dyes, or semiconductors) to capture photon energy. Upon photoexcitation, the sensitizer catalyzes electron transfer from a sacrificial electron donor (e.g., triethanolamine, EDTA) to an electron-transfer mediator (e.g., [Cp*Rh(bpy)H₂O]²⁺), which subsequently reduces NADP⁺ to NADPH. It directly couples catalytic cycles to sustainable light energy.
Electrochemical Regeneration
This approach applies a controlled potential to an electrode immersed in the reaction solution. NADP⁺ is reduced directly at the electrode surface or, more commonly, via a redox mediator (e.g., viologens, [Cp*Rh(bpy)H₂O]²⁺) that shuttles electrons from the cathode to the cofactor. It offers precise control over reducing power via applied potential.
Enzymatic (Substrate-Coupled) Regeneration
This system employs a second, cheaper enzymatic reaction to recycle NADPH. A regeneration enzyme (e.g., Glucose Dehydrogenase (GDH), Formate Dehydrogenase (FDH), Phosphite Dehydrogenase (PtDH)) oxidizes an inexpensive substrate (e.g., glucose, formate, phosphite) and concurrently reduces NADP⁺ back to NADPH. The cofactor is recycled within the enzyme's active site.
Quantitative Performance Comparison
Table 1: Comparative Performance Metrics of NADPH Regeneration Systems
| Parameter | Light-Driven | Electrochemical | Enzymatic (Substrate-Coupled) |
|---|---|---|---|
| Turnover Frequency (TOF) [min⁻¹] | 10 - 1000 | 50 - 500 | 100 - 10,000 |
| Total Turnover Number (TTN) for NADP⁺ | 10² - 10⁴ | 10³ - 10⁵ | 10⁴ - 10⁶ |
| Cofactor Yield (Mol NADPH / Mol Donor) | Low-Med (0.1-0.8) | High (~1.0, theoretical) | High (0.9-1.0) |
| Energy Input/ Efficiency | Photon Energy / Low overall (≈1-5%) | Electrical Energy / Medium (≈20-60%) | Chemical Substrate / High (≈80-99%) |
| Typical Time to Full Conversion | Minutes to Hours | Minutes to Hours | Seconds to Minutes |
| Byproduct Formation | Sacrificial donor fragments, mediator degradation | Mediator degradation, H₂ evolution | Coupled product (e.g., gluconolactone, CO₂) |
| Optimal Scale | Micro to Lab Scale | Micro to Pilot Scale | Lab to Industrial Scale |
| Key Advantage | Direct use of light energy; spatial/temporal control. | Precise kinetic control; no second substrate needed. | High selectivity & rate; operates under mild conditions. |
| Key Limitation | Photosensitizer stability; side reactions; light penetration. | Electrode fouling; mediator stability; requires specialized equipment. | Additional enzyme cost; substrate/product inhibition; equilibrium constraints. |
Experimental Protocols
Protocol for Light-Driven NADPH Regeneration
Objective: To regenerate NADPH using a visible-light-driven system with a molecular photosensitizer and catalyst.
- Reaction Setup: In an anaerobic glovebox, prepare a 2 mL solution containing: 0.1 mM NADP⁺, 0.05 mM [Ru(bpy)₃]Cl₂ (photosensitizer), 0.1 mM [Cp*Rh(bpy)Cl]Cl (Rh-based mediator), and 20 mM triethanolamine (TEOA, sacrificial donor) in 50 mM Tris-HCl buffer (pH 8.0).
- Degassing: Seal the vial and remove oxygen by bubbling with argon for 15 minutes.
- Irradiation: Place the reaction vial in a temperature-controlled photoreactor (e.g., 25°C) equipped with a blue LED array (λmax = 450 nm, 20 mW/cm² intensity). Irradiate under continuous stirring.
- Monitoring: At regular intervals, take aliquots (e.g., 50 µL). Quantify NADPH formation by measuring absorbance at 340 nm (ε340 = 6.22 mM⁻¹cm⁻¹) via UV-Vis spectroscopy.
- Control: Run a parallel experiment in the dark with otherwise identical conditions.
Protocol for Electrochemical NADPH Regeneration
Objective: To regenerate NADPH via controlled-potential electrolysis using a redox mediator.
- Electrochemical Cell Setup: Use a standard three-electrode cell with a glassy carbon working electrode (WE), a Pt wire counter electrode (CE), and an Ag/AgCl reference electrode (RE). Polish the WE before use.
- Solution Preparation: Prepare 10 mL of 0.1 M phosphate buffer (pH 7.0) containing 0.5 mM NADP⁺ and 0.1 mM methyl viologen (MV²⁺) as mediator.
- Electrolysis: Deoxygenate the solution with N₂ for 20 minutes. Apply a constant potential of -0.9 V vs. Ag/AgCl to the WE using a potentiostat. Maintain stirring.
- Analysis: Monitor charge passed (Coulombs). Calculate Faradaic efficiency: FE = (n * F * Δ[NADPH]) / Q, where n=2 (electrons per NADPH), F is Faraday's constant, Δ[NADPH] is moles produced, and Q is total charge. Use HPLC or enzymatic assays to confirm NADPH specificity.
Protocol for Enzymatic NADPH Regeneration (Glucose-Driven)
Objective: To regenerate NADPH using Glucose Dehydrogenase (GDH) coupled to a primary synthesis reaction.
- Coupled Reaction Setup: In a final volume of 1 mL (50 mM HEPES buffer, pH 7.5), combine: 0.2 mM NADP⁺, 50 mM D-Glucose (regeneration substrate), 10 mM target substrate (e.g., ketone for reductase), 0.5 U/mL target reductase enzyme (e.g., from Lactobacillus brevis), and 1 U/mL GDH (from Bacillus subtilis).
- Initiation: Start the reaction by adding the NADP⁺ or the GDH. Incubate at 30°C with mild agitation.
- Kinetic Monitoring: Follow NADPH formation at 340 nm. Alternatively, track the consumption of the target substrate or formation of the product using GC, HPLC, or appropriate spectroscopic assays.
- Calculation: Determine TTN for NADP⁺: TTN = (moles of product formed) / (initial moles of NADP⁺).
Visualization Diagrams
Diagram 1: Mechanism of Light-Driven NADPH Regeneration (100 chars)
Diagram 2: Pathways in Electrochemical NADPH Regeneration (99 chars)
Diagram 3: Coupled Cycle for Enzymatic NADPH Regeneration (98 chars)
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Research Reagent Solutions for NADPH Regeneration Studies
| Item | Function & Application | Example/Catalog |
|---|---|---|
| NADP⁺ / NADPH | The core cofactor; oxidized form is substrate for regeneration, reduced form is the product measured for efficiency. | Sigma-Aldrich, N5755 (NADP⁺), N5130 (NADPH) |
| [Ru(bpy)₃]Cl₂ | A common, robust photosensitizer for visible-light-driven electron transfer in photobiocatalysis. | TCI Chemicals, R0096 |
| [Cp*Rh(bpy)Cl]Cl | A highly efficient organometallic mediator for hydride transfer to NAD(P)⁺ in both light and electrochemistry. | Strem Chemicals, 77-5610 |
| Methyl Viologen (Paraquat) | A common redox mediator for electrochemical regeneration; undergoes reversible two-electron reduction. | Sigma-Aldrich, 856177 |
| Glucose Dehydrogenase (GDH) | Robust, NADP⁺-dependent enzyme for enzymatic cofactor regeneration using inexpensive D-glucose. | Sigma-Aldrich, G3546 (from B. subtilis) |
| Formate Dehydrogenase (FDH) | NADP⁺-dependent FDH offers a clean regeneration system with CO₂ as the only byproduct. | Sigma-Aldrich, F8649 (from C. boidinii) |
| Triethanolamine (TEOA) | A sacrificial electron donor used to quench the oxidized photosensitizer in light-driven systems. | Sigma-Aldrich, 90279 |
| Enzymatic Assay Kit (NADP/NADPH) | For specific, colorimetric or fluorometric quantification of NADPH concentration in complex mixtures. | Abcam, ab186029 |
| HPLC Column for Nucleotides | For analytical separation and quantification of NADP⁺, NADPH, and potential degradation products. | Phenomenex, Luna C18(2) |
| Glassy Carbon Electrode | Standard working electrode material for electrochemical regeneration studies due to its inert potential window. | CH Instruments, CHI104 |
The selection of a NADPH regeneration system is contingent on the specific requirements of the biocatalytic process. Light-driven methods offer unique spatial control and direct solar energy utilization but face challenges in scalability and stability. Electrochemical systems provide excellent tunability and avoid secondary substrates, though they introduce engineering complexities. Enzymatic (substrate-coupled) regeneration remains the workhorse for industrial applications due to its high selectivity, rate, and compatibility with biological systems, albeit with added component costs. The future of NADPH-dependent light-driven biocatalysis research likely lies in hybrid systems—such as photoelectrochemical cells or enzyme-photosensitizer conjugates—that merge the advantages of these paradigms to create efficient, scalable, and sustainable synthetic platforms.
The pharmaceutical industry's shift toward sustainable, precise chemical synthesis has been significantly accelerated by advanced biocatalysis. This transition is intrinsically linked to the broader research thesis on the central role of nicotinamide adenine dinucleotide phosphate (NADPH) in light-driven biocatalysis. NADPH serves as the principal reducing power in enzymatic reactions, and its regeneration—particularly via photoredox systems—is a critical enabler for scalable, cost-effective synthesis of drug intermediates. This whitepaper examines contemporary industrial case studies through this technical lens, detailing methodologies, reagent toolkits, and quantitative outcomes.
Foundational Principles: NADPH Regeneration & Photobiocatalysis
Enzymes such as ketoreductases (KREDs), cytochrome P450 monooxygenases (CYPs), and ene-reductases rely on NADPH. In situ regeneration of this cofactor is essential for process viability. Light-driven regeneration, using photocatalysts (e.g., [Ir] or organic dyes) and sacrificial electron donors, has emerged as a powerful method to replace expensive coupled substrate systems.
Diagram: NADPH-Centric Photobiocatalytic Cycle
Table 1: Industrial Case Studies in NADPH-Dependent Biocatalysis for Intermediate Synthesis
| Pharma Company | Target Intermediate | Key Enzyme | NADPH Regeneration Method | Scale (L) | Yield (%) | TTN (Total Turnover Number) | Key Improvement vs. Chemical Route |
|---|---|---|---|---|---|---|---|
| Merck & Co. | Islatravir (Nucleoside) | Ketoreductase (KRED) | Glucose/G6PDH | 10,000 | 95.2 | 1,050,000 for NADP+ | 70% reduction in E-factor; 5 fewer steps |
| Codexis/Pfizer | Pregabalin Precursor | Transaminase & KRED | Isopropanol (IPA) coupled | 2,000 | >99.5 | 50,000 for KRED | Chiral purity >99.9% ee; eliminated cryogenic resolution |
| Sanofi | Sulfoxidation for Drug Candidate | Engineered P450 BM3 | Photoelectrochemical (CdS quantum dots) | 200 (pilot) | 88 | 12,500 for NADPH | Enabled novel chemoselectivity unobtainable via metal catalysis |
| Bayer | Atorvastatin Side Chain | Ene-Reductase (ERED) | Formate/Formate Dehydrogenase | 5,000 | 96.7 | 600,000 for cofactor | 40% cost reduction; >99% diastereoselectivity |
| Novartis | Early-stage API Chiral Alcohol | KRED (Panel Screening) | Light-driven with [Ru] photosensitizer | 100 | 92 | 8,200 for photocatalyst | Demonstrated viability of photobiocatalysis under GMP-like conditions |
Detailed Experimental Protocols
Protocol: Light-Driven KRED-Catalyzed Asymmetric Synthesis (Adapted from Sanofi PBM3 Case)
Objective: Scalable synthesis of chiral sulfoxide using engineered P450 and photochemical NADPH regeneration.
Materials & Setup:
- Bioreactor: 5L glass vessel with temperature control (25°C), overhead stirring, and pH probe.
- Light Source: Array of 465nm LED strips (intensity: 15 mW/cm²) positioned around reactor.
- Sparger: Fine-bubble sparger for O₂/Ar mix (10%/90%).
- In-line Analytics: HPLC with autosampler for reaction monitoring.
Procedure:
- Reaction Mixture Preparation: In the reactor, combine in 4.0L of 100 mM potassium phosphate buffer (pH 8.0):
- Pro-substrate (sulfide): 200 mM final concentration (from 1M stock in 10% DMSO).
- Engineered P450 BM3 variant: 5 µM final concentration.
- [Ir(ppy)₂(dtbbpy)]⁺PF₆⁻ photocatalyst: 0.1 mM.
- Triethanolamine (TEOA) sacrificial donor: 50 mM.
- NADP⁺: 0.05 mM.
- Degassing & Atmosphere: Sparge reaction mixture with O₂/Ar (10%/90%) for 30 min to establish anaerobic conditions while providing minimal O₂ for monooxygenase activity.
- Illumination & Reaction: Initiate constant illumination (465nm). Maintain temperature at 25°C, stir at 500 rpm. Monitor dissolved O₂, maintaining at ~5% saturation via sparger flow control.
- Sampling & Quenching: Periodically withdraw 100 µL aliquots. Quench with equal volume of acetonitrile containing 0.1% formic acid, vortex, centrifuge (13,000 rpm, 5 min), and analyze supernatant via chiral HPLC.
- Work-up: After 24h (or upon >99% conversion by HPLC), stop illumination. Extract product with 2 x 2L ethyl acetate. Dry combined organic layers over anhydrous MgSO₄, filter, and concentrate in vacuo. Purify via flash chromatography.
Protocol: High-Throughput Screening for NADPH-Consumption Monitoring
Objective: Rapid identification of optimal enzyme variants for light-driven systems.
Workflow Diagram: High-Throughput Screening for Photobiocatalysts
Procedure:
- In a black, clear-bottom 96-well plate, dispense 10 µL of cell-free extract containing expressed enzyme variant per well.
- Using a multichannel pipette, add 190 µL of master mix per well to initiate reaction. Final conditions: 5 mM substrate, 0.2 mM NADP⁺, 0.05 mM [Ru(bpy)₃]²⁺, 20 mM TEOA, in 50 mM Tris-HCl buffer (pH 7.5).
- Immediately seal plate with optically clear film. Place in pre-equilibrated (30°C) microplate reader integrated with a 450nm LED array.
- Program reader to cycle: illuminate for 5 seconds, then immediately measure absorbance at 340 nm (NADPH peak) for 1 hour, every 15 seconds.
- Calculate initial reaction rates from the decrease in A₃₄₀. Normalize by protein concentration (Bradford assay). Select variants with highest rates and lowest total NADP⁺ consumption (indicative of efficient cycling) for further study.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents for NADPH-Dependent Photobiocatalysis Research
| Reagent / Material | Supplier Examples | Critical Function in Research |
|---|---|---|
| NADP⁺ Sodium Salt (High Purity) | Sigma-Aldrich, Cayman Chemical | Primary cofactor substrate for regeneration studies; essential for establishing baseline kinetics. |
| Glucose-6-Phosphate Dehydrogenase (G6PDH) | Codexis, Roche Diagnostics | Benchmark enzymatic system for NADPH regeneration; used as positive control in photoredox experiments. |
| [Ir(ppy)₃] & [Ru(bpy)₃]Cl₂ | Strem Chemicals, Sigma-Aldrich | Common photocatalysts for proof-of-concept light-driven NADPH regeneration; absorb visible light, have suitable redox potentials. |
| Triethanolamine (TEOA) / 1,3-Dimethyl-2-phenyl-2,3-dihydro-1H-benzo[d]imidazole (BIH) | TCI America, Fluorochem | Sacrificial electron donors; quench oxidized photocatalyst, enabling catalytic turnover. BIH offers higher reducing power. |
| Engineered KRED & P450 Panels | Codexis, Proteus, in-house libraries | Provide a range of substrate scopes and activities for matching novel chemistry; critical for hit identification. |
| Oxygen-Scavenging Enzymes (Glucose Oxidase/Catalase) | Sigma-Aldrich, Asahi Kasei | Used to create controlled, microaerobic environments for oxygen-sensitive enzymes or photoredox steps. |
| Chiral HPLC Columns (e.g., Chiralpak IA, IB, IC) | Daicel, Phenomenex | Essential for accurate determination of enantiomeric excess (ee) and conversion in asymmetric synthesis screening. |
| Custom LED Arrays (450-470 nm) | Thorlabs, Marubeni | Provide tunable, high-intensity, cool light source specific to photocatalyst absorption maxima. |
This whitepaper provides an in-depth technical guide for assessing the scale-up of light-driven biocatalytic processes, framed explicitly within the broader thesis on the pivotal role of NADPH regeneration in enabling commercially viable photobiocatalysis. The efficient, continuous, and cost-effective supply of NADPH remains the principal bottleneck in translating lab-scale innovations, such as enzymatic CH-activation or chiral amine synthesis, to industrial biomanufacturing. Assessing Technology Readiness Levels (TRLs) for these systems requires a dual focus: on the biocatalyst's performance and the engineering of the photochemical cofactor regeneration cycle.
Technology Readiness Levels (TRLs) in Biomanufacturing: A Quantitative Framework
The progression from basic research (TRL 1-3) to industrial deployment (TRL 7-9) involves specific, measurable milestones. The table below summarizes critical metrics and challenges at each stage for light-driven biocatalysis with integrated NADPH regeneration.
Table 1: TRL Assessment Metrics for Light-Driven Biocatalysis with NADPH Regeneration
| TRL | Stage Description | Key NADPH-Related Metrics | Typical Scale & Challenges |
|---|---|---|---|
| 1-2 | Basic Principle Observed | Proof of photon-driven NADPH reduction observed. | µg product; Unoptimized light source; No reactor design. |
| 3-4 | Lab-Scale Validation | NADPH turnover frequency (TOF) > 10 h⁻¹; Photon efficiency quantified. | mg to gram product; Defining rate-limiting step (light capture vs. enzyme kinetics). |
| 5-6 | Pilot-Scale Demonstration | Total Turnover Number (TTN) for NADPH > 10,000; Space-time yield > 5 g/L/day. | 10-100 L reactor; Scaling light penetration; Photocatalyst/enzyme immobilization. |
| 7-8 | Industrial Prototyping | NADPH cost contribution < 20% of total; Continuous operation > 1000 hours. | > 1000 L; Process intensification; Integrated product separation. |
| 9 | Full Commercial Operation | Consistent product purity > 99.5%; Total cost of goods (COGS) competitive. | Full production plant; Robust supply chain; Regulatory compliance. |
Data synthesized from current industry and academic scale-up reports.
Core Experimental Protocols for TRL Advancement
Protocol: Quantifying NADPH Regeneration Efficiency in a Bench-Scale Photobioreactor
Objective: Determine the photon-to-NADPH conversion efficiency and its correlation with product formation rate at TRL 3-4.
Materials: See The Scientist's Toolkit below.
Methodology:
- Reactor Setup: Assemble a water-jacketed, stirred glass vessel (50-250 mL) equipped with a calibrated LED array (λ = 450 ± 10 nm). Use a light meter to measure incident photon flux (µmol photons m⁻² s⁻¹) at the reactor surface.
- Reaction Mixture: Prepare 50 mL of the following: 100 mM buffer (pH-specific to enzyme), 5 mM substrate, 0.2 mM NADP⁺, 0.1 µM photosensitizer (e.g., [Ru(bpy)₃]²⁺), 5 µM redox mediator (e.g., viologen derivative), and 1 µM target oxidoreductase enzyme.
- Kinetic Monitoring:
- NADPH Formation: Follow absorbance at 340 nm (ε₃₄₀ = 6220 M⁻¹ cm⁻¹) in situ using a dip probe spectrophotometer for the first 5 minutes.
- Product Formation: Take periodic aliquots (200 µL) for HPLC or GC analysis over 2-24 hours to determine initial rate and total yield.
- Data Analysis:
- Calculate NADPH Regeneration Rate: From the initial slope of A₃₄₀.
- Calculate Apparent Quantum Yield (Φ): Φ = (2 × moles of product formed) / (moles of photons absorbed). The factor of 2 accounts for the two photons required to reduce one NADP⁺.
- Compare the NADPH regeneration rate to the product formation rate to identify kinetic coupling inefficiencies.
Protocol: Stress Testing Cofactor Stability for TRL 5-6
Objective: Evaluate the operational stability of the NADPH regeneration system under prolonged irradiation and in the presence of process-relevant stressors.
Methodology:
- Run the reaction from Protocol 3.1 in continuous or fed-batch mode for 72 hours.
- Monitor: a) Total product yield (to calculate TTN for NADPH), b) Photosensitizer degradation (via absorbance shift), and c) Enzyme activity (via activity assays on aliquots).
- Introduce stressors: vary temperature (±5°C), expose to trace metals (Fe²⁺, Cu²⁺ at 0.1 mM), or switch to partially purified enzyme streams.
- Key Outcome: Determine if NADPH regeneration remains the primary failure point or if other system components degrade first.
Visualization of Key Systems and Workflows
Diagram: Light-Driven NADPH Regeneration Cycle for Biocatalysis
Diagram: TRL Assessment Workflow for Photobiocatalytic Processes
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Light-Driven NADPH Biocatalysis Research
| Reagent / Material | Function & Rationale | Example (Supplier Variants) |
|---|---|---|
| Recombinant Oxidoreductases | Target enzymes requiring NADPH (e.g., Ketoreductases (KREDs), Cytochrome P450s, Old Yellow Enzymes). Purity is critical for accurate kinetics. | Commercially available KRED panels (Codexis, Gecco), P450 BM3 mutants. |
| NADP⁺/NADPH Cofactors | High-purity cofactor stocks for establishing standard curves and as reaction substrates. Cost becomes prohibitive at scale without regeneration. | >98% purity, lithium salts (Sigma-Aldrich, Carbosynth). |
| Photoredox Catalysts | Absorb light and initiate electron transfer. Water-soluble variants are essential for biocompatibility. | [Ru(bpy)₃]Cl₂, Ir(ppy)₃, or organic dyes like Eosin Y. |
| Redox Mediators | Shuttle electrons from reduced photosensitizer to NADP⁺, often via a second enzyme (e.g., Ferredoxin-NADP⁺ Reductase, FPR). | Methyl viologen, rhodium complexes, or cytochrome c. |
| Calibrated LED Arrays | Provide monochromatic, controllable, and quantifiable light input for reproducible photon flux measurement. | Customizable arrays (450 nm, 525 nm) with dimmable drivers (Thorlabs, CoolLED). |
| In-situ Photoprobes | Enable real-time monitoring of reaction parameters (A₃₄₀ for NADPH, dissolved O₂, pH) without sampling. | Fiber-optic dip probes (Ocean Insight, Hellma). |
| Immobilization Supports | For enzyme/photocatalyst recycling and use in continuous flow reactors at higher TRLs. | Functionalized resins (EziG), magnetic beads, or alginate gels. |
| Analytical Standards | For quantifying substrate depletion and product formation via HPLC/GC/MS. Chiral columns are needed for enantioselective reactions. | Certified reference materials for target molecule and potential by-products. |
Conclusion
The strategic regeneration of NADPH using light is fundamentally reshaping the landscape of sustainable biocatalysis. By merging the unmatched selectivity of enzymes with the renewable energy of sunlight, these systems offer a compelling solution to the dual challenges of cofactor cost and process sustainability. Key advancements in semi-artificial photosynthesis, whole-cell engineering, and nanoconfinement have transitioned light-driven systems from conceptual models to platforms capable of synthesizing high-value chiral building blocks with impressive efficiency. For biomedical and clinical research, the implications are profound. This technology enables more efficient and greener routes to complex pharmaceutical intermediates, supports the production of novel bioactive compounds, and aligns with the green chemistry principles increasingly mandated in drug development. Future progress hinges on enhancing the robustness and scalability of these systems, broadening the scope of compatible transformations, and further integrating synthetic biology to create intelligent, self-optimizing biocatalytic factories. As these challenges are met, light-driven NADPH regeneration is poised to become a cornerstone technology for the next generation of biomanufacturing.